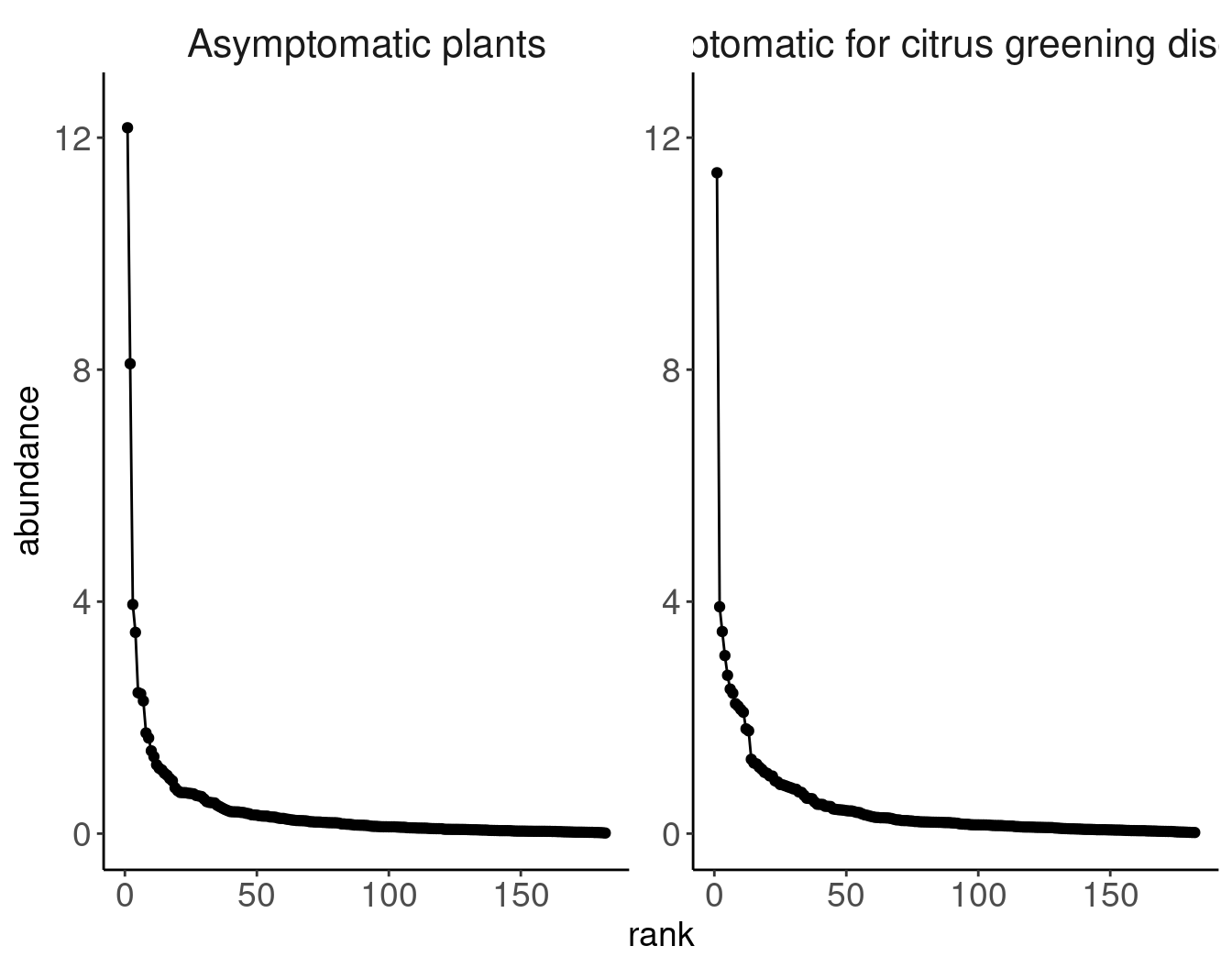
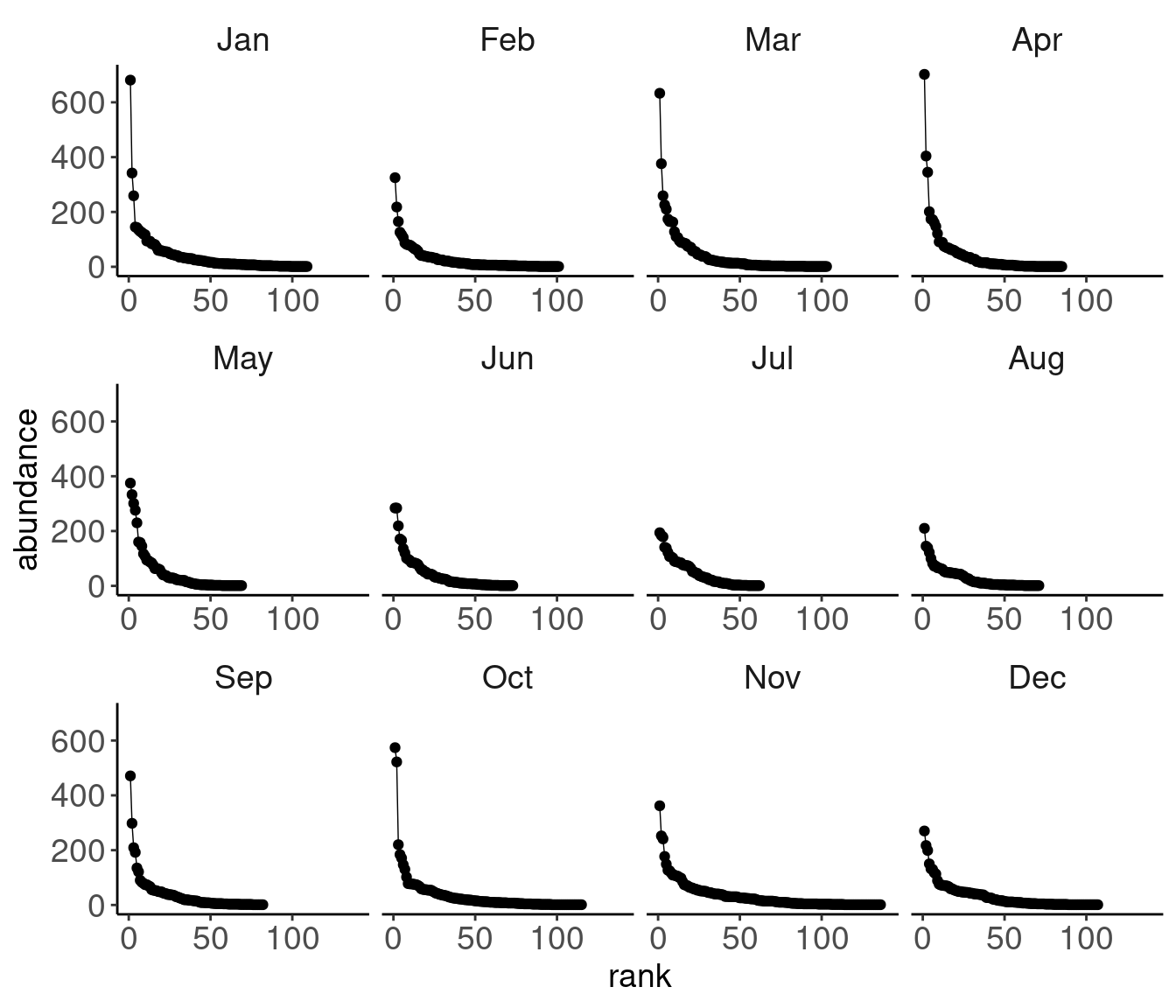
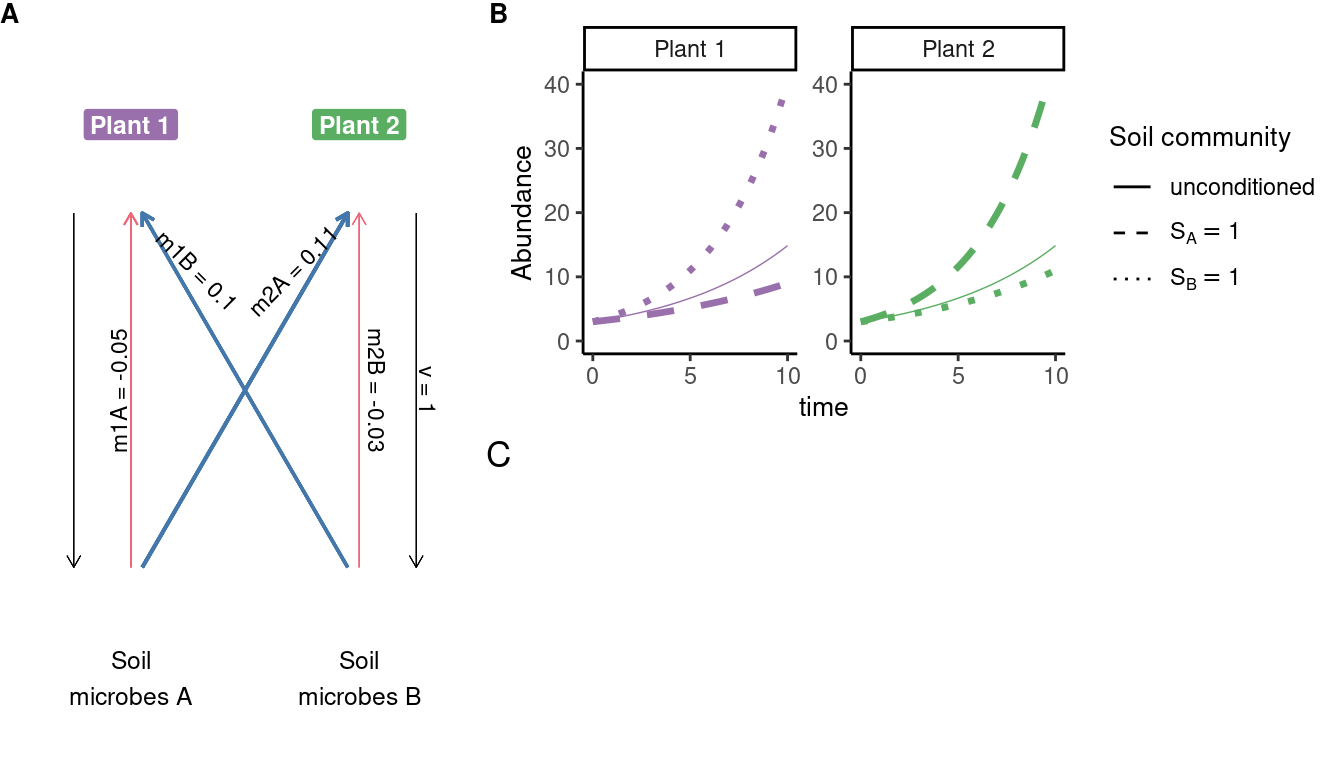
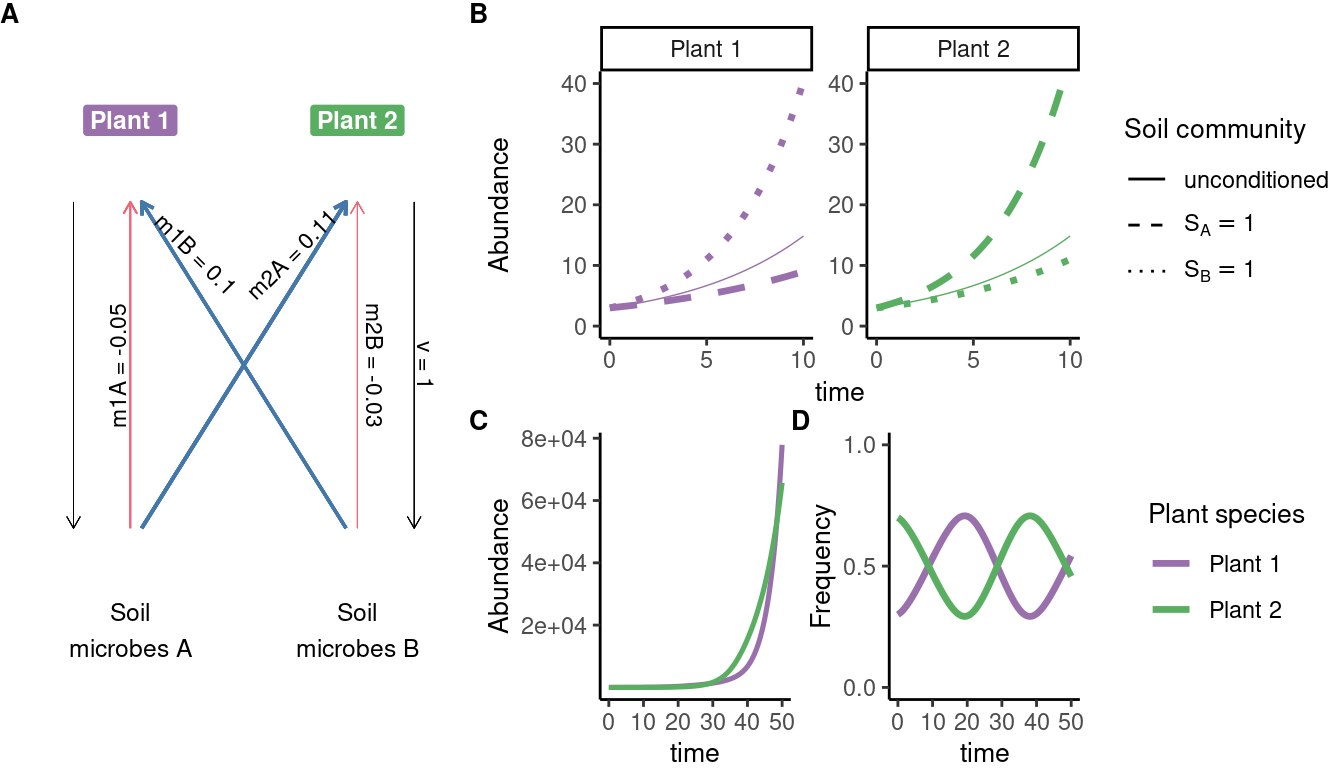
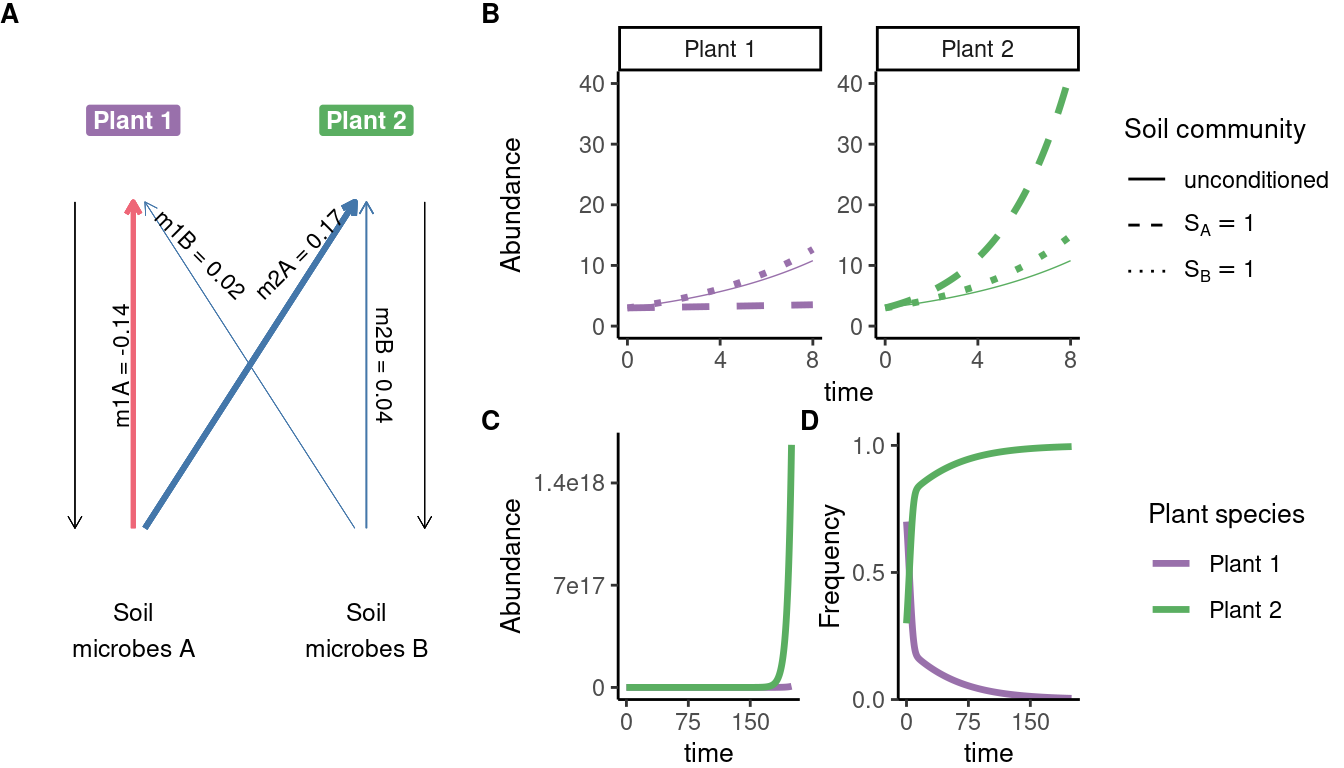
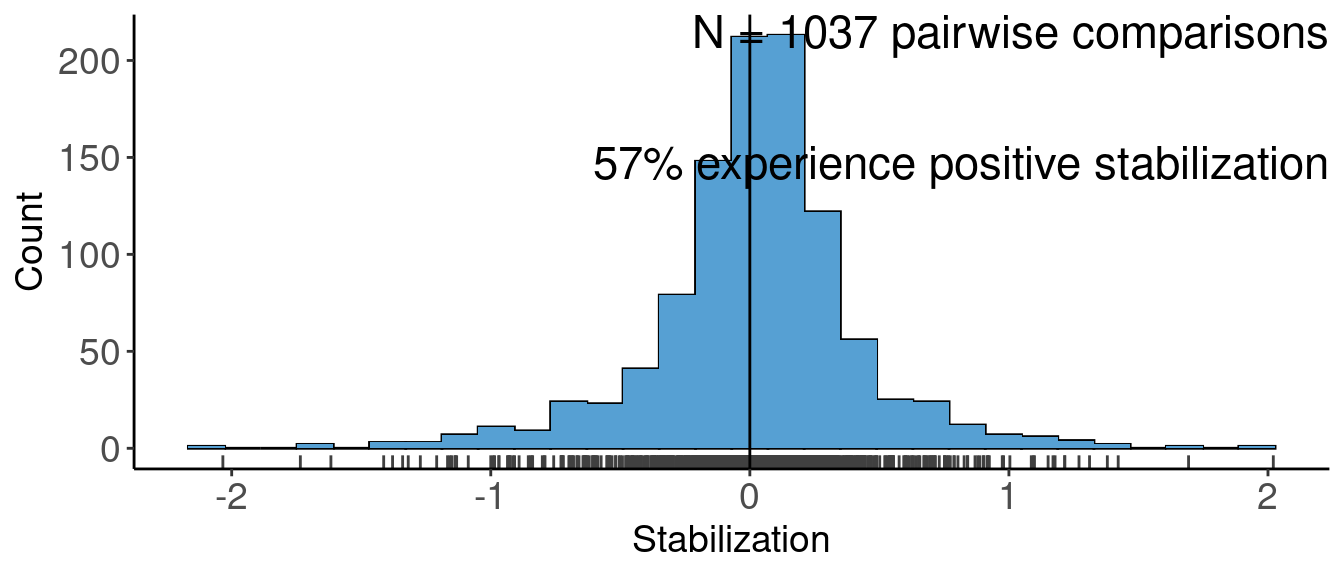
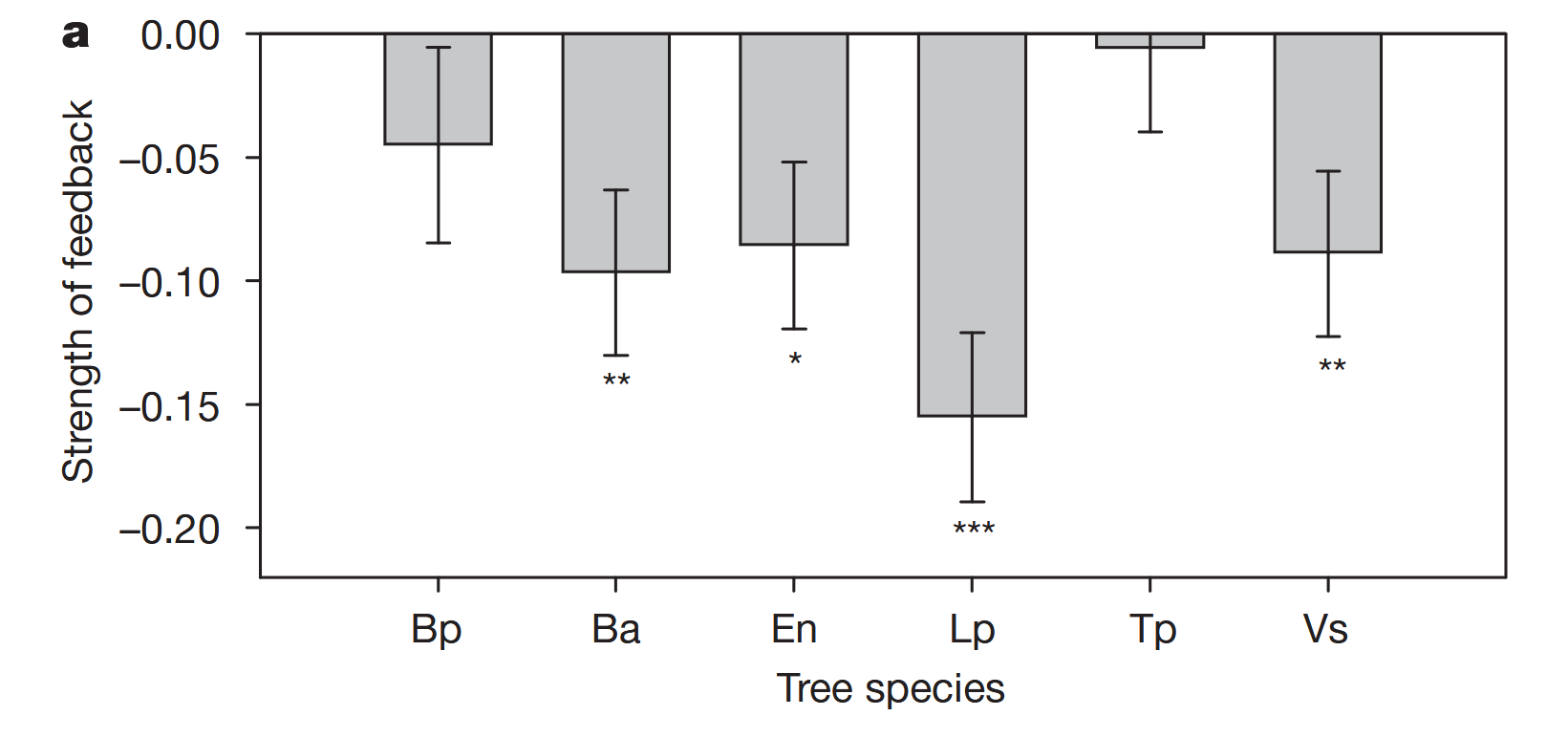
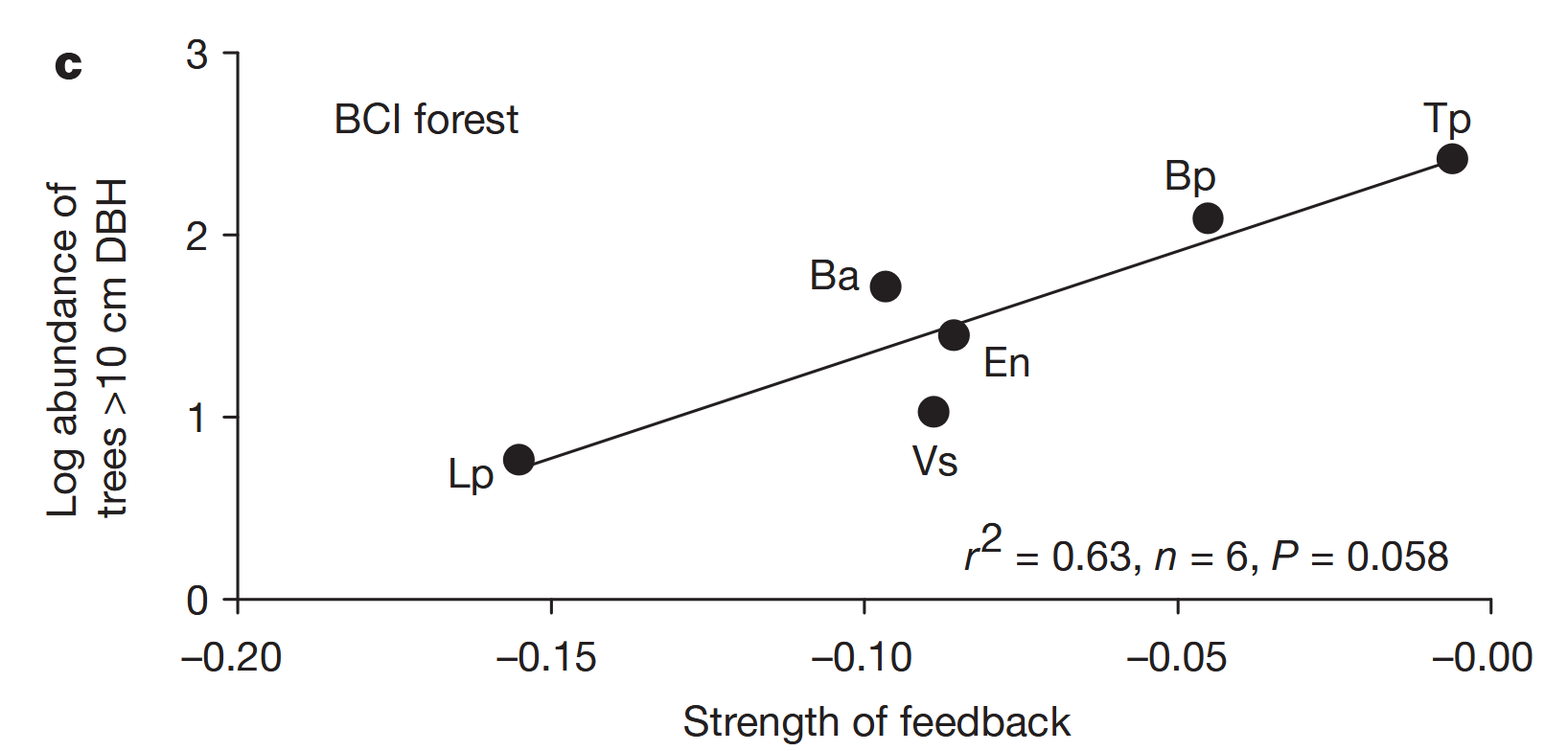
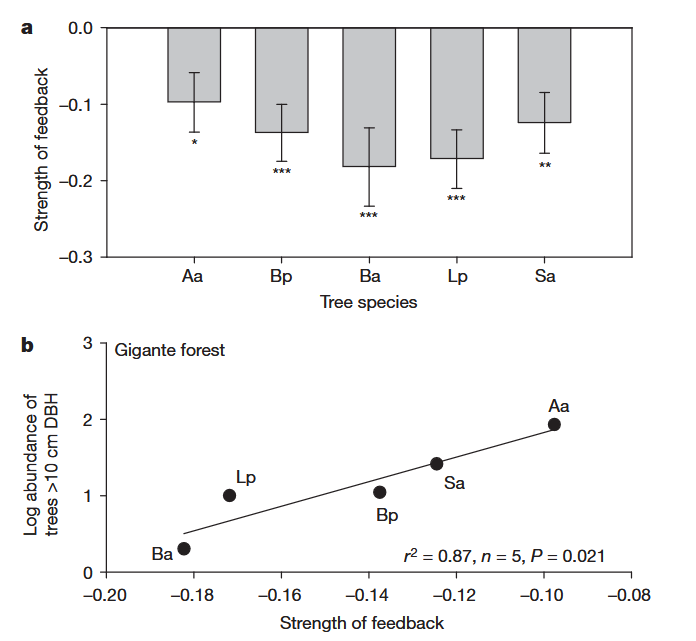
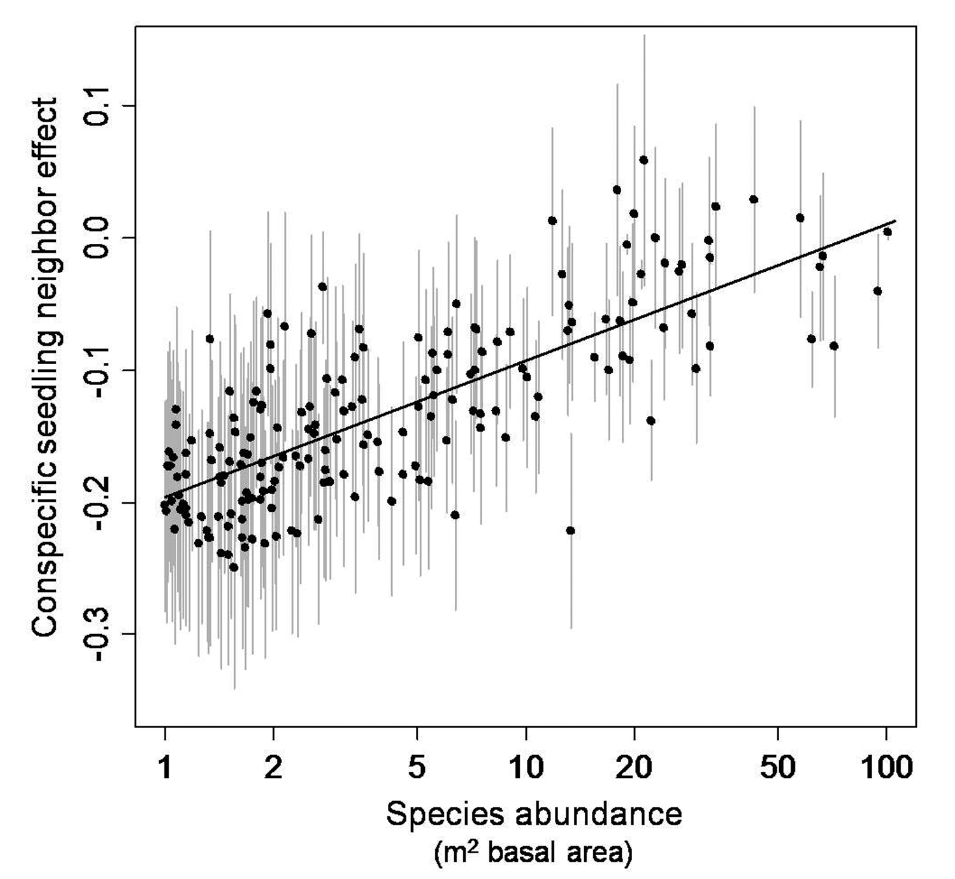
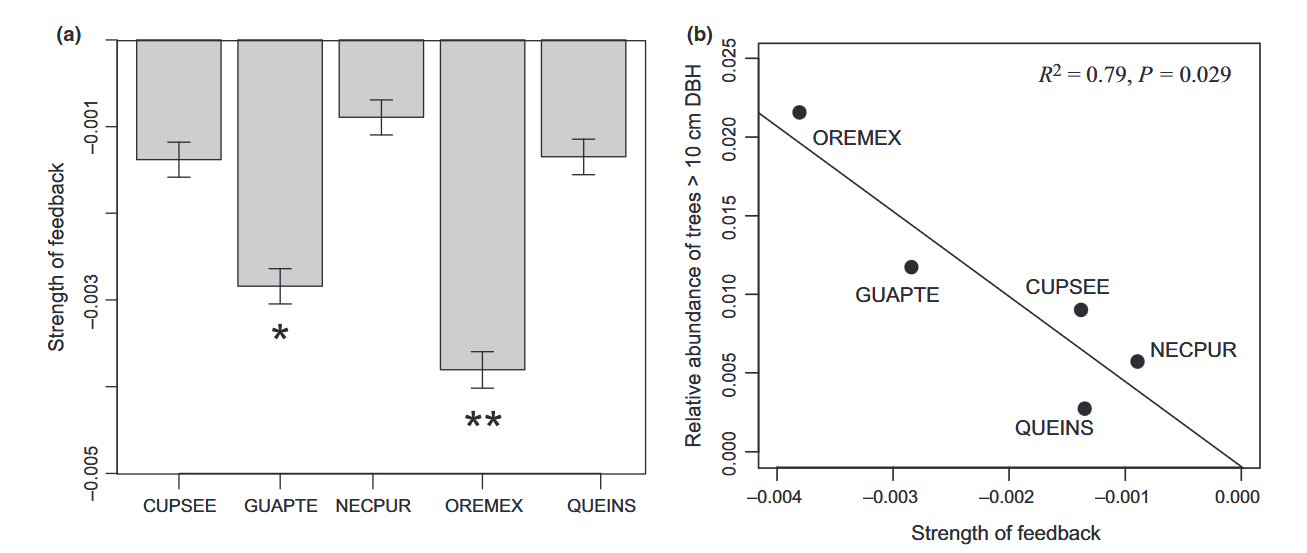
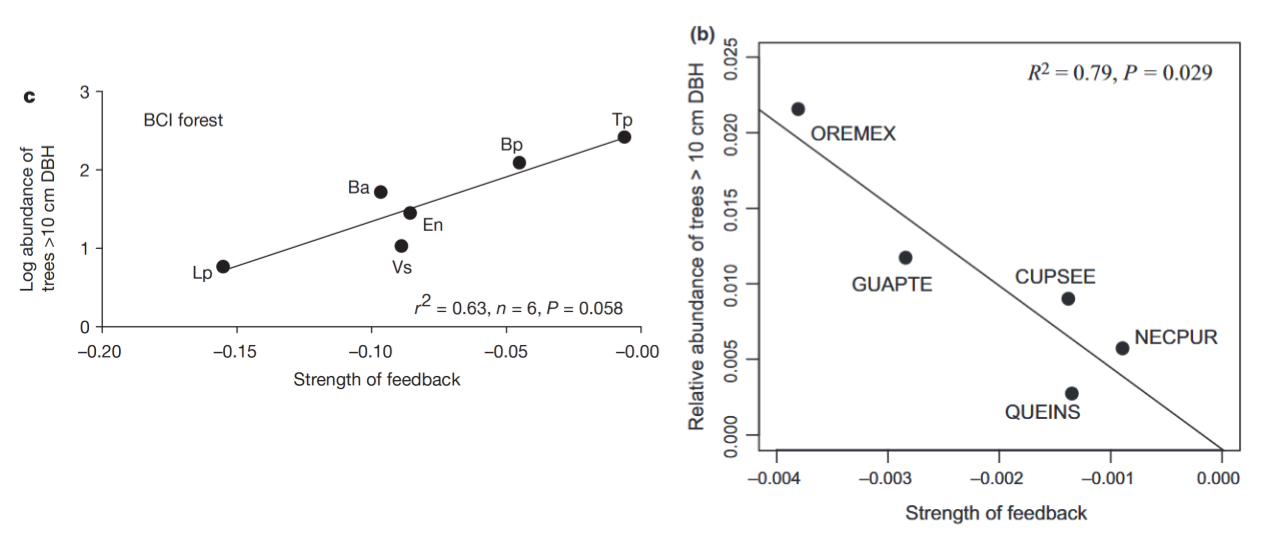
# data extracted from Fig. 3 of Klironomos 2002, https://www.nature.com/articles/417067a.pdf
# using WebPlotDigitizer
tribble(
~fb, ~abun,
-0.34, 3.27,
-0.41, 7.94,
-0.27, 2.20,
0.01, 41.49,
-0.14, 27.39,
-0.05, 67.29,
0.00, 59.28,
-0.16, 7.81,
0.11, 35.77,
-0.08, 42.83,
-0.15, 1.63,
-0.36, 1.22,
-0.07, 1.33,
-0.27, 5.30,
-0.40, 2.27,
0.06, 87.34,
-0.21, 4.75,
-0.23, 0.38,
-0.02, 35.58,
-0.05, 41.78,
-0.25, 1.67,
-0.20, 7.32,
-0.15, 12.71,
-0.09, 10.36,
-0.18, 11.18,
0.04, 53.85,
-0.29, 6.85,
-0.14, 31.00,
-0.33, 3.01,
-0.07, 15.24,
-0.15, 67.60,
0.00, 14.95,
-0.22, 28.98,
0.11, 50.46,
-0.03, 51.82,
-0.23, 18.16,
-0.15, 0.34,
-0.36, 0.44,
-0.30, 1.19,
0.20, 62.78,
-0.30, 7.89,
-0.14, 21.72,
-0.37, 5.60,
-0.13, 64.76,
-0.26, 0.91,
-0.16, 45.70,
-0.24, 6.83,
-0.43, 2.80,
-0.13, 29.19,
-0.49, 0.77,
-0.05, 13.94,
-0.06, 37.92,
0.01, 68.30,
-0.36, 10.76,
-0.31, 7.38,
-0.27, 4.26,
0.06, 60.02,
0.04, 71.89,
-0.23, 0.63,
-0.01, 80.93,
) |>
ggplot(aes(x = fb, y = abun)) +
geom_point(size = 2.5) +
# fit line from Fig 3 of the paper
geom_function(fun = function(x) 114.529*x^2 + 156.652*x + 44.013) +
xlab("Species-level feedback") +
ylab("Relative abundance")