Belowground regulation of
aboveground dynamics
Gaurav Kandlikar
Louisiana State University, Baton Rouge
slides: https://talks.gklab.org/tulane-25
Sal (Shorea) dominated forests in northern India, Photo by Dhritiman Mukherjee
Intercropping of soybean and wheat; image from U. Iowa
“Christmas tree scientists work to manage grinchy fungal foes”; image from Washington State Univ
Research goals
Explain patterns of diversity in nature
Predict responses to perturbations
Approach


Approach


Today’s talk
Ecological theory for microbial effects on plant coexistence
Microbial effects on plant communities in variable environments
Mathematical underpinning of plant–soil feedback
See also Bever, Westover, and Antonovics (1997) and Kandlikar (2024)
Plant dynamics
\[\frac{dN_1}{dt} = w_1N_1 ~~~~ \text{and} ~~~ \frac{dN_2}{dt} = w_2N_2\]
\[w_i = m_{iA}f_A + m_{iB}(1-f_A)\]
where \(f_i\) is the relative frequency of microbial community \(i\) in the soil (e.g. \(f_A = \frac{N_A}{N_A+N_B}\))
Interpretation: Plants have exponential population growth at a rate determined by the composition of the soil community.
Microbe dynamics
\[\frac{dN_A}{dt} = N_A\frac{N_1}{N_1 + N_2}~~~\text{and}~~~\frac{dN_B}{dt} = \nu N_B\frac{N_2}{N_1 + N_2}\]
Interpretation: Soil communities grow at a rate determined by the frequency of each plant (and the relative strength of each plant’s conditioning effect, \(\nu\)).
R code to simulate model dynamics
Code
psf_model <- function(time, init, params) {
with (as.list(c(time, init, params)), {
# description of parameters (see Bever et al. 1997)
# N1 and N2: abundance of the plant species 1 and 2
# p1: frequency of plant species 1; p2 = 1-pA
# m1A, m1B, m2A, m2B: conspecific and heterospecific effects of microbial community A or B on the growth of plant 1 or 2
# pA: frequency of the soil microbial community A
# v: influence of plant species 2 on the microbial community relative to that of plant 1
# Differential equations
dN1 <- (m1A*pA + m1B*(1-pA))*N1
dN2 <- (m2A*pA + m2B*(1-pA))*N2
dp1 <- p1*(1-p1)*((m1A-m2A)*pA + (m1B-m2B)*(1-pA))
dpA <- pA*(1-pA)*(p1-v*(1-p1))
# Return dN1 and dN2
return(list(c(dN1, dN2, dp1, dpA)))
})
}\[ \overbrace{(m_{1B} + m_{2A})}^{\text{interspecific effects}}- \overbrace{(m_{1A} + m_{2B})}^{\text{intraspecific effects}} > 0 \]
\[ \underbrace{\frac{1}{2}\big[\overbrace{(m_{1B} + m_{2A})}^{\text{interspecific effects}}- \overbrace{(m_{1A} + m_{2B})}^{\text{intraspecific effects}}\big]}_{\text{Stablization}} > 0 \]
Lasting influence of the Bever model
Code
# Dataset downloaded from Figshare:
# curl::curl_download("https://figshare.com/ndownloader/files/14874749", destfile = "../crawford-supplement.xlsx")
read_excel("../data/crawford-supplement.xlsx", sheet = "Data") |>
mutate(stab = -0.5*rrIs) |> # show in terms of stabilization instead of I_s
filter(stab > -3) |> # Remove the pair with Stab approx. 6 -- according to McCarthy Neuman (original author), this is likely due to bad seed survival
ggplot(aes(x = stab)) +
geom_histogram(color = "black") +
geom_histogram(fill = "#56A0D3") +
geom_rug(color = "grey25") +
geom_vline(xintercept = 0) +
annotate("text", x = Inf, y = Inf, label = "N = 1037 pairwise comparisons\n
57% experience positive stabilization",
vjust=1, hjust = 1, size = 6) +
xlab("Stabilization") + ylab("Count")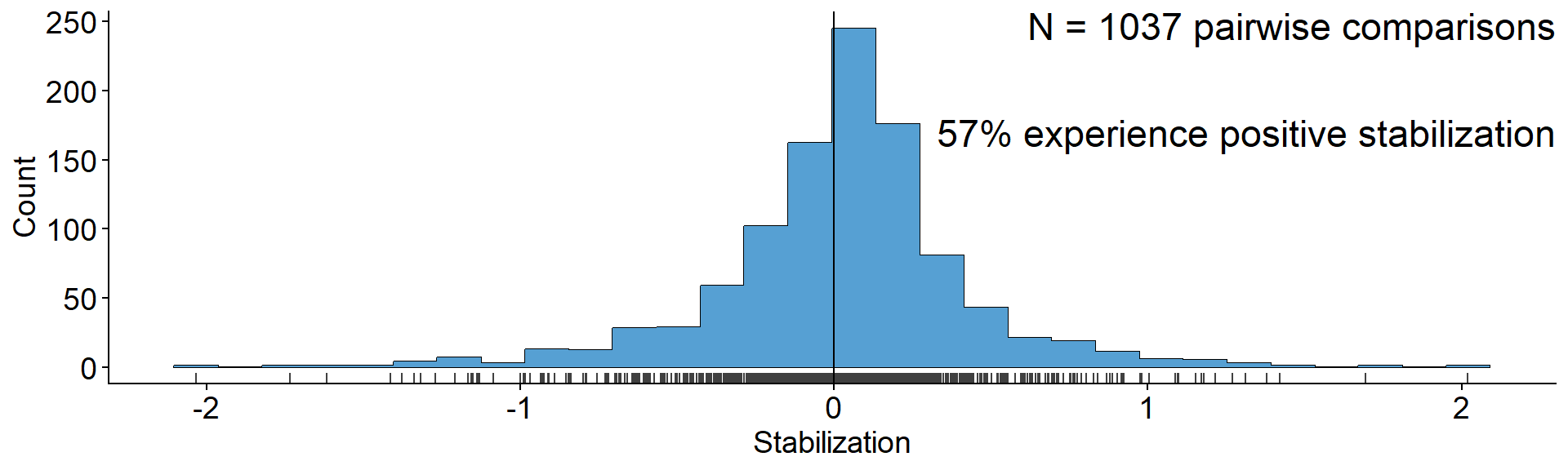
Data from Crawford et al. (2019)
A puzzle: same stabilization, different model dynamics
Code
# Define a function used for making the PSF framework
# schematic, given a set of parameter values
make_params_plot <- function(params, scale = 1.5) {
color_func <- function(x) {
ifelse(x < 0, "#EE6677", "#4477AA")
}
df <- data.frame(x = c(0,0,1,1),
y = c(0,1,0,1),
type = c("M", "P", "M", "P"))
params_plot <-
ggplot(df) +
annotate("text", x = 0, y = 1.1, size = 3.25, label = "Plant 1", color = "white", fill = "#9970ab", fontface = "bold") +
annotate("text", x = 1, y = 1.1, size = 3.25, label = "Plant 2", color = "white", fill = "#5aae61", fontface = "bold") +
annotate("text", x = 0, y = -0.15, size = 3.25, label = "Soil\nmicrobes A", label.size = 0, fill = "transparent") +
annotate("text", x = 1, y = -0.15, size = 3.25, label = "Soil\nmicrobes B", label.size = 0, fill = "transparent") +
annotate("text", x = 0.45, y = -0.4, size = 3.5, fill = "transparent",
label.size = 0.05,
label = (paste0("Stabilization = ",
0.5*(params["m1B"] + params["m2A"] - params["m1A"] - params["m2B"])))) +
geom_segment(aes(x = 0, xend = 0, y = 0.1, yend = 0.9),
arrow = arrow(length = unit(0.03, "npc")),
linewidth = abs(params["m1A"])*scale,
color = alpha(color_func(params["m1A"]), 1)) +
geom_segment(aes(x = 0.05, xend = 0.95, y = 0.1, yend = 0.9),
arrow = arrow(length = unit(0.03, "npc")),
linewidth = abs(params["m2A"])*scale,
color = alpha(color_func(params["m1B"]),1)) +
geom_segment(aes(x = 0.95, xend = 0.05, y = 0.1, yend = 0.9),
arrow = arrow(length = unit(0.03, "npc")),
linewidth = abs(params["m1B"])*scale,
color = alpha(color_func(params["m2A"]), 1)) +
geom_segment(aes(x = 1, xend = 1, y = 0.1, yend = 0.9),
arrow = arrow(length = unit(0.03, "npc"),),
linewidth = abs(params["m2B"])*scale,
color = alpha(color_func(params["m2B"]), 1)) +
# Plant cultivation of microbes
geom_segment(aes(x = -0.25, xend = -0.25, y = 0.9, yend = 0.1), linewidth = 0.15, linetype = 1,
arrow = arrow(length = unit(0.03, "npc"))) +
geom_segment(aes(x = 1.25, xend = 1.25, y = 0.9, yend = 0.1), linewidth = 0.15, linetype = 1,
arrow = arrow(length = unit(0.03, "npc"))) +
annotate("text", x = 0, y = 0.5,
label = (paste0("m1A = ", params["m1A"])),
angle = 90, vjust = -0.25, size = 3) +
annotate("text", x = 1.15, y = 0.5,
label = (paste0("m2B = ", params["m2B"])),
angle = -90, vjust = 1.5, size = 3) +
annotate("text", x = 0.75, y = 0.75,
label = (paste0("m2A = ", params["m2A"])),
angle = 45, vjust = -0.25, size = 3) +
annotate("text", x = 0.25, y = 0.75,
label = (paste0("m1B = ", params["m1B"])),
angle = -45, vjust = -0.25, size = 3) +
xlim(c(-0.4, 1.4)) +
coord_cartesian(ylim = c(-0.25, 1.15), clip = "off") +
theme_void() +
theme(legend.position = "none",
plot.caption = element_text(hjust = 0.5, size = 10))
return(params_plot)
}
# Define a function for simulating the dynamics of the
# PSF model with deSolve
params_coex <- c(m10 = 0.16, m20 = 0.16,
m1A = 0.11, m1B = 0.26,
m2A = 0.27, m2B = 0.13, v = 1)
time <- seq(0,50,0.1)
init_pA_05 <- c(N1 = 3, N2 = 7, p1 = 0.3, pA = 0.3)
out_pA_05 <- ode(y = init_pA_05, times = time, func = psf_model, parms = params_coex) |> data.frame()
params_to_plot_coex <- c(m1A = unname(params_coex["m1A"]-params_coex["m10"]),
m1B = unname(params_coex["m1B"]-params_coex["m10"]),
m2A = unname(params_coex["m2A"]-params_coex["m20"]),
m2B = unname(params_coex["m2B"]-params_coex["m20"]), v=1)
param_plot_coex <-
make_params_plot(params_to_plot_coex, scale = 6) +
annotate("text", x = 1.375, y = 0.5,
label = paste0("v = ", params_coex["v"]),
angle = -90, vjust = 1.5, size = 3)
panel_abund_coex <-
out_pA_05 |>
as_tibble() |>
select(time, N1, N2) |>
pivot_longer(N1:N2) |>
ggplot(aes(x = time, y = value, color = name)) +
geom_line(linewidth = 0.9) +
scale_color_manual(values = c("#9970ab", "#5aae61"),
name = "Plant species", label = c("Plant 1", "Plant 2"), guide = "none") +
scale_y_continuous(breaks = c(2e4, 4e4, 6e4, 8e4), labels = scales::scientific) +
ylab("Abundance") +
theme(axis.title = element_text(size = 10))
# Panel D: plot for frequencies of both plants when growing
# in dynamic soils
panel_freq_coex <-
out_pA_05 |>
as_tibble() |>
mutate(p2 = 1-p1) |>
select(time, p1, p2) |>
pivot_longer(p1:p2) |>
ggplot(aes(x = time, y = value, color = name)) +
geom_line(linewidth = 1.2) +
scale_color_manual(values = c("#9970ab", "#5aae61"),
name = "Plant species", label = c("Plant 1", "Plant 2")) +
scale_y_continuous(limits = c(0,1), breaks = c(0, 0.5, 1)) +
ylab("Frequency") +
theme(axis.title = element_text(size = 10))
param_plot_coex + {panel_abund_coex+panel_freq_coex &
theme(axis.text = element_text(size = 8), legend.position = 'none')} +
plot_layout(widths = c(1/3, 2/3))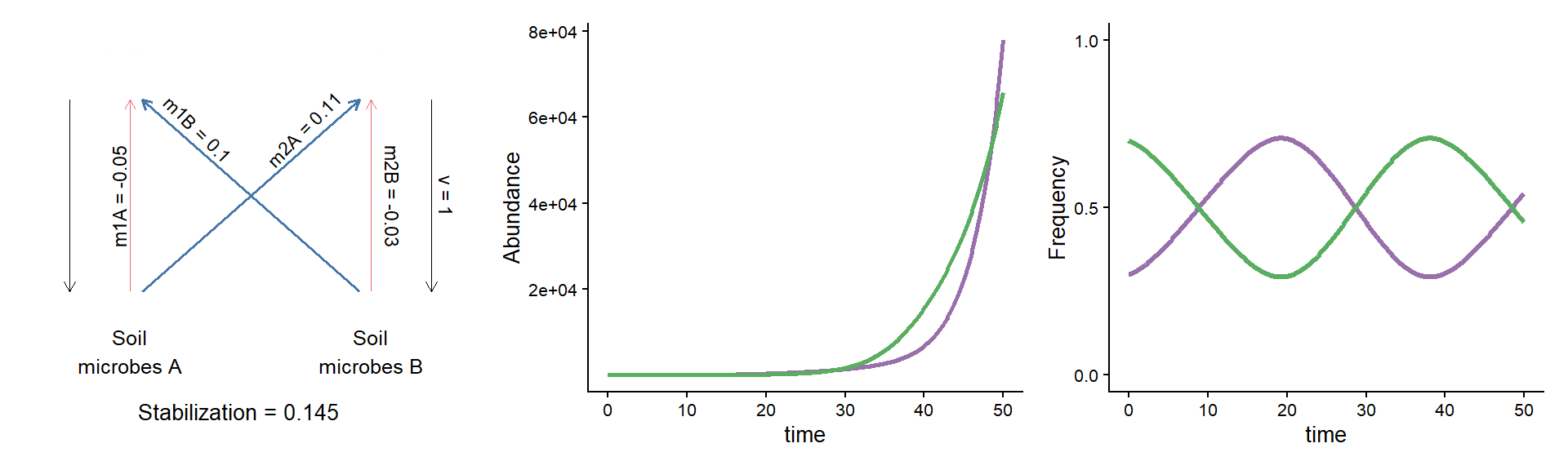
Code
# Define a function for simulating the dynamics of the
# PSF model with deSolve
params <- c(m10 = 0.16, m20 = 0.16,
m1A = 0.02, m2A = 0.33,
m1B = 0.18, m2B = 0.20, v = 1)
time <- seq(0,200,0.1)
init_pA_05 <- c(N1 = 3, N2 = 7, p1 = 0.3, pA = 0.3)
out_pA_05 <- ode(y = init_pA_05, times = time, func = psf_model, parms = params) |> data.frame()
params_to_plot <- c(m1A = unname(params["m1A"]-params["m10"]),
m1B = unname(params["m1B"]-params["m10"]),
m2A = unname(params["m2A"]-params["m20"]),
m2B = unname(params["m2B"]-params["m20"]), v=1)
param_plot <-
make_params_plot(params_to_plot, scale = 6) +
annotate("text", x = 1.375, y = 0.5,
label = paste0("v = ", params["v"]),
angle = -90, vjust = 1.5, size = 3)
panel_abund <-
out_pA_05 |>
as_tibble() |>
select(time, N1, N2) |>
pivot_longer(N1:N2) |>
ggplot(aes(x = time, y = value, color = name)) +
geom_line(linewidth = 0.9) +
scale_color_manual(values = c("#9970ab", "#5aae61"),
name = "Plant species", label = c("Plant 1", "Plant 2"), guide = "none") +
scale_y_continuous(breaks = c(0,8e17,1.6e18), labels = c("0", "7e17","1.4e18")) +
ylab("Abundance")
# Panel D: plot for frequencies of both plants when growing
# in dynamic soils
panel_freq <-
out_pA_05 |>
as_tibble() |>
mutate(p2 = 1-p1) |>
select(time, p1, p2) |>
pivot_longer(p1:p2) |>
ggplot(aes(x = time, y = value, color = name)) +
geom_line(linewidth = 1.2) +
scale_color_manual(values = c("#9970ab", "#5aae61"),
name = "Plant species", label = c("Plant 1", "Plant 2")) +
scale_y_continuous(limits = c(0,1), breaks = c(0, 0.5, 1)) +
ylab("Frequency")
param_plot + {panel_abund + panel_freq &
theme(axis.text = element_text(size = 8), legend.position = 'none')} +
plot_layout(widths = c(1/3, 2/3))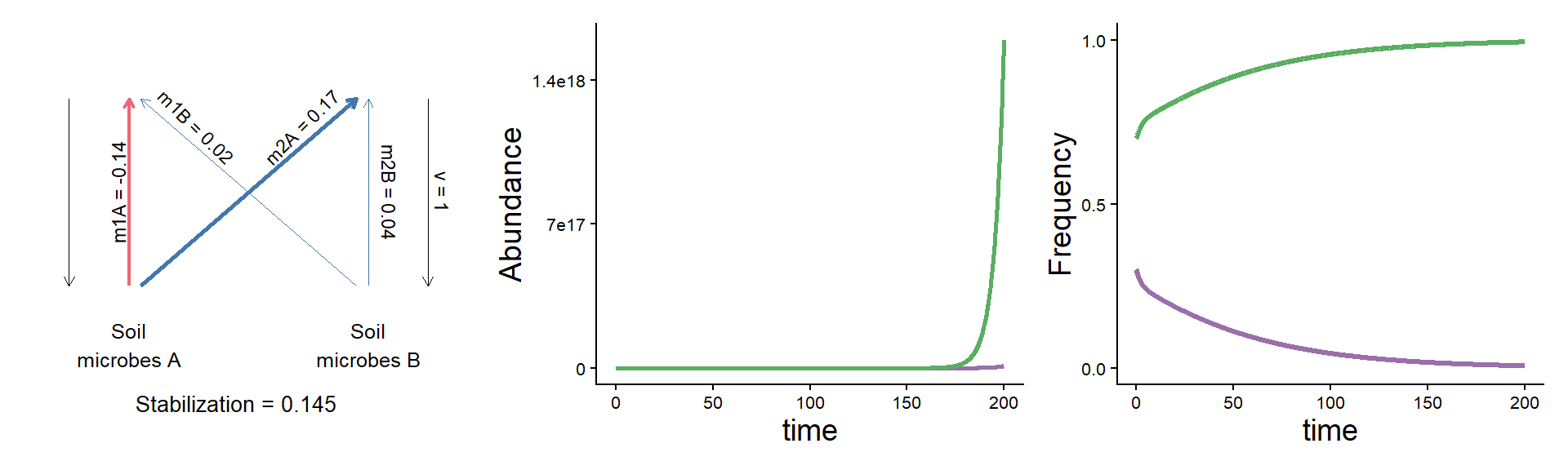
How can we more accurately infer predicted plant dynamics under plant–soil feedback?
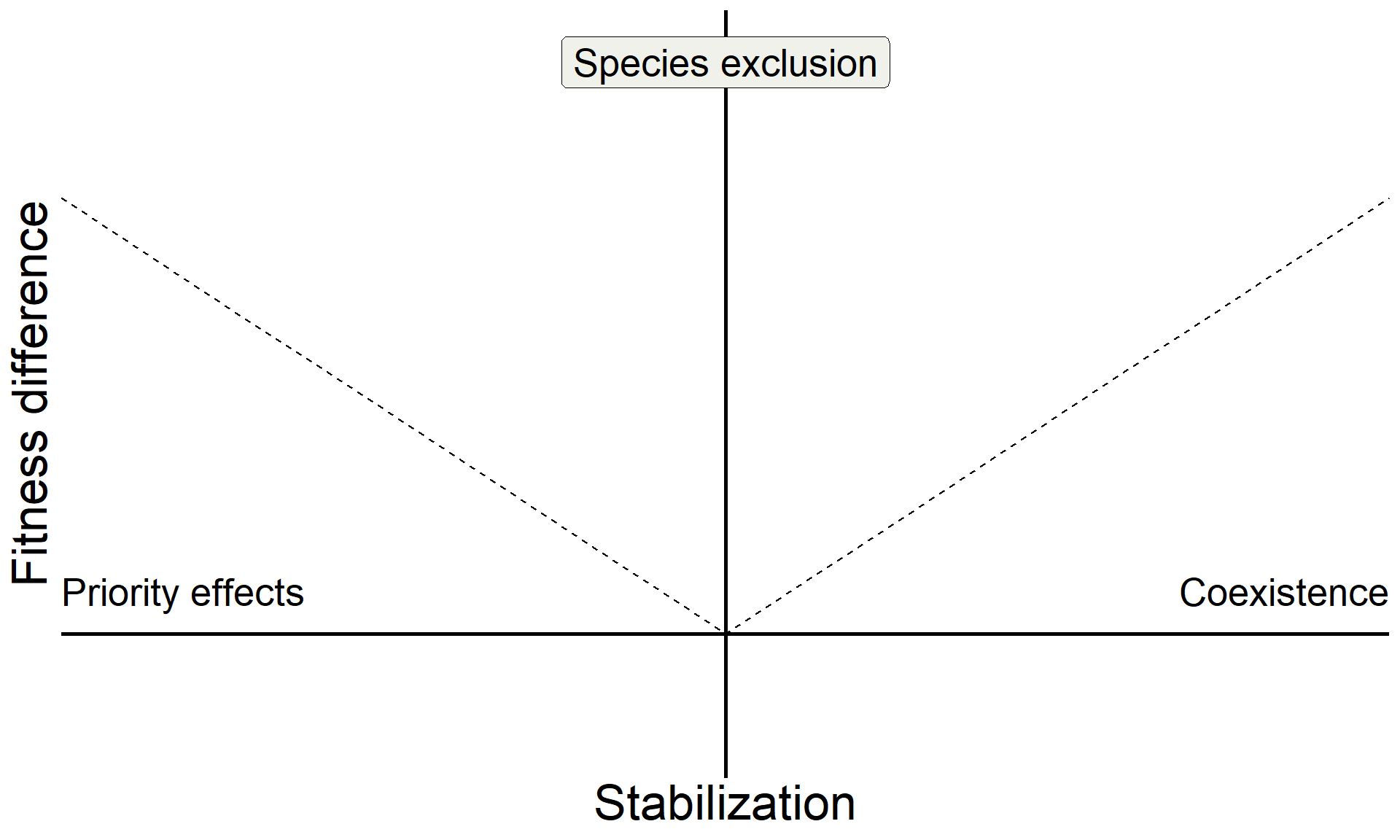
Build on insights from coexistence theory (Chesson 2018):
- Stablization is necessary for coexistence but doesn’t guarantee coexistence
- Stable coexistence is possible only when stabilization overcomes competitive imbalances (fitness differences)

How can we more accurately infer predicted plant dynamics under plant–soil feedback?
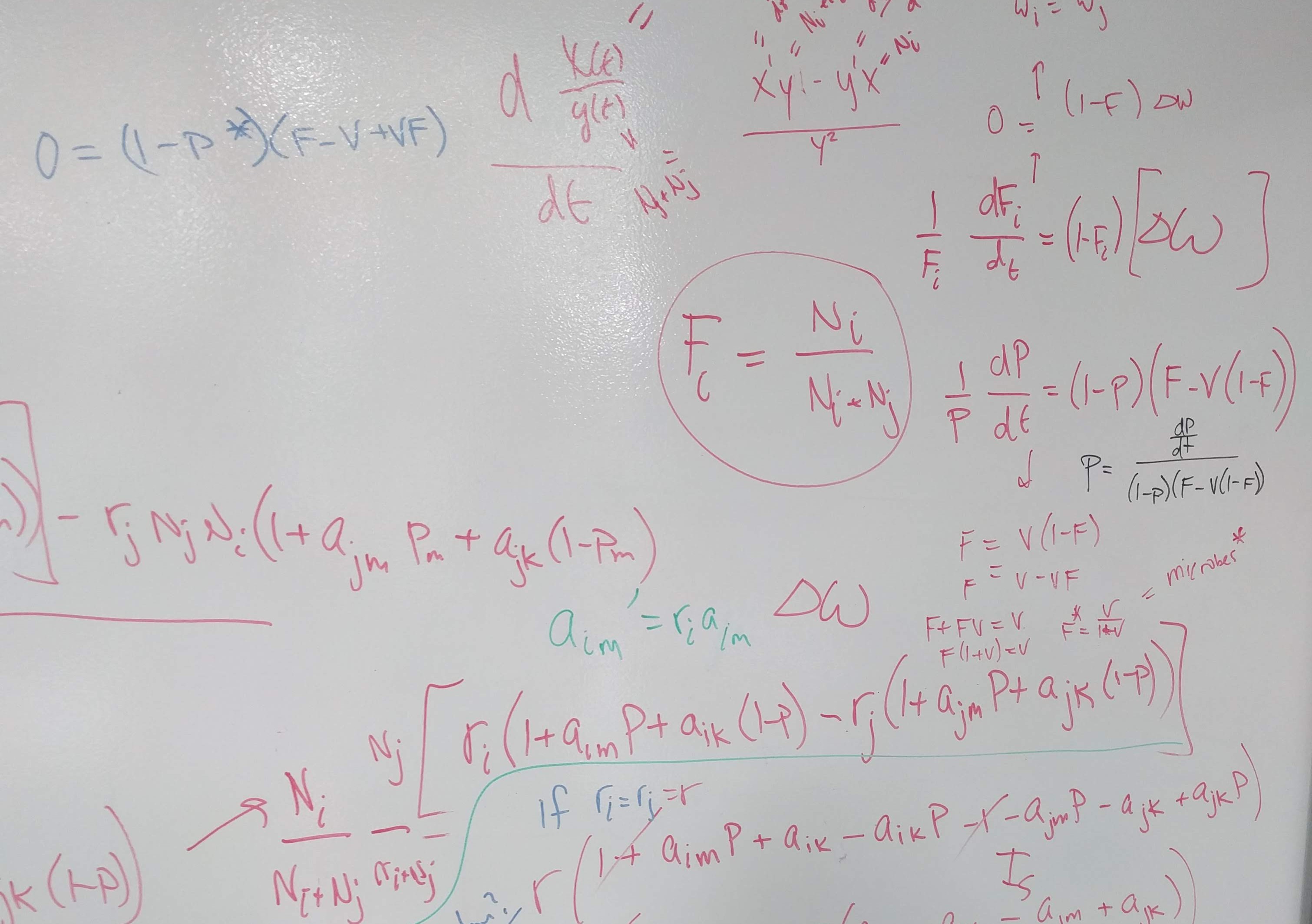
How can we more accurately infer predicted plant dynamics under plant–soil feedback?

How can we more accurately infer predicted plant dynamics under plant–soil feedback?

How can we more accurately infer predicted plant dynamics under plant–soil feedback?

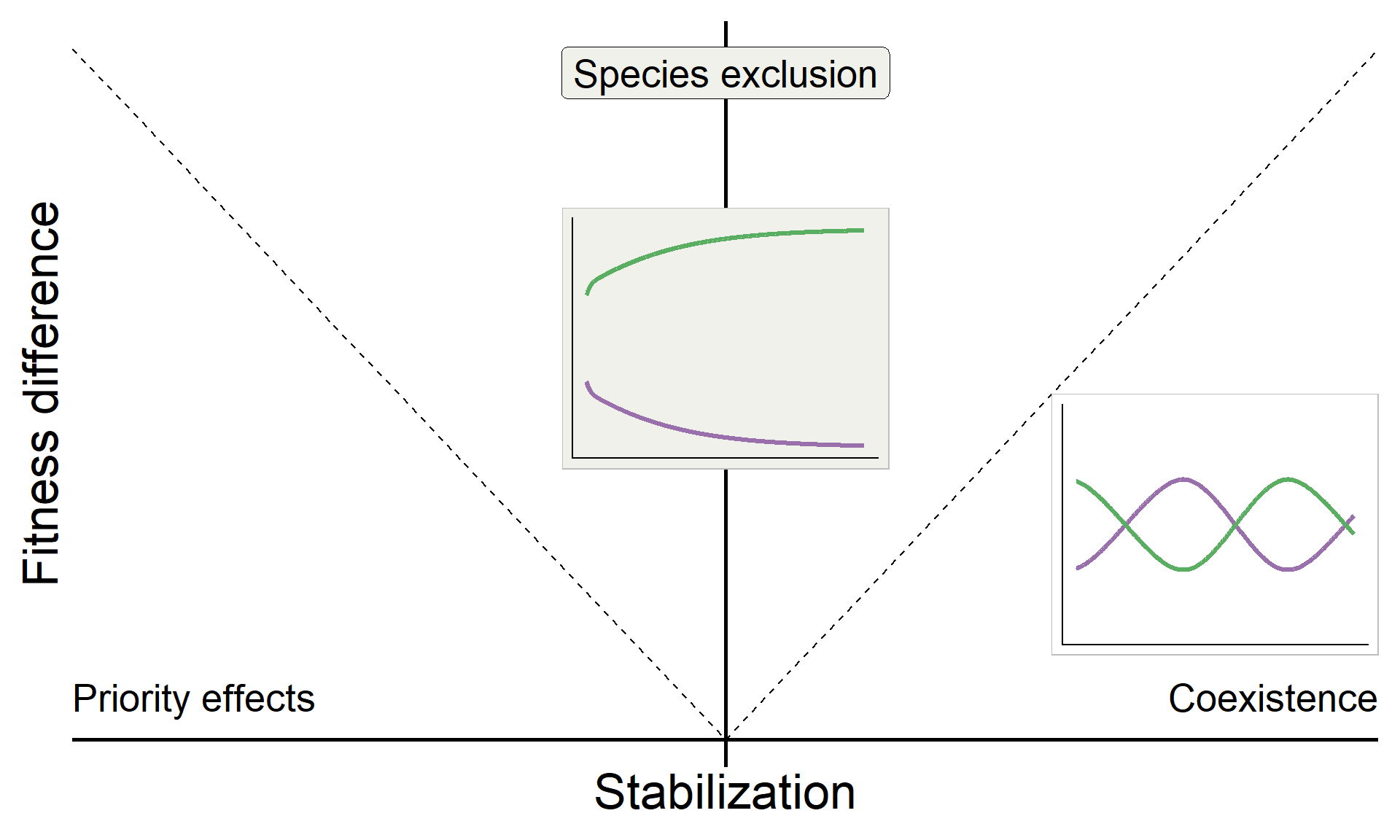
\[\text{Fitness difference}_{1,2} = \overbrace{\frac{1}{2}(m_{1A}+m_{1B})}^{\substack{\text{Sensitivity of}\\ \text{Sp 1 to microbes}}} - \overbrace{\frac{1}{2}(m_{2A}+m_{2B})}^{\substack{\text{Sensitivity of}\\ \text{Sp 2 to microbes}}}\]
Code
base <-
tibble(sd = seq(-1,1,0.1),
fd = c(seq(1,0,-0.1), seq(0.1,1,0.1))) |>
ggplot(aes(x = sd, y = fd)) +
geom_line(linetype = "dashed") +
geom_hline(yintercept = 0, linewidth = 1) +
geom_vline(xintercept = 0, linewidth = 1) +
xlab("Stabilization") +
ylab("Fitness difference") +
theme(axis.text = element_blank(),
axis.ticks = element_blank(),
axis.line = element_blank(),
axis.title = element_text(size = 25)) +
scale_x_continuous(expand = c(0,0)) +
scale_y_continuous(expand = c(0.04,0))
base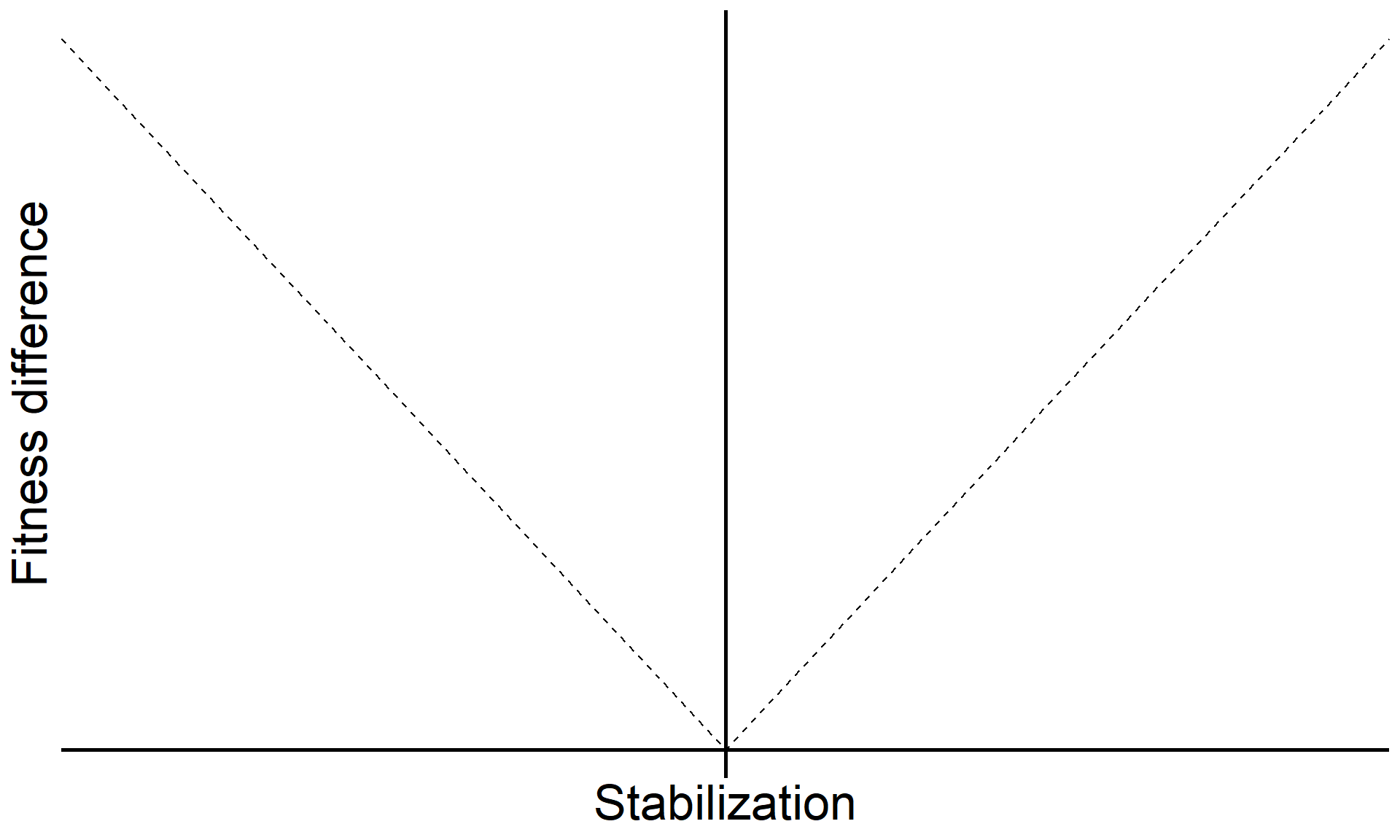
Code
base <-
base +
annotate("text", x = Inf, y = 0, hjust = 1, vjust = -1, label = "Coexistence", size = 7, family = "italic") +
annotate("text", x = -Inf, y = 0, hjust = 0, vjust = -1, label = "Priority effects", size = 7, family = "italic") +
annotate("label", x = 0, y = Inf, vjust = 1.5, label = "Species exclusion", size = 7, fill = "#F0F1EB",family = "italic")
base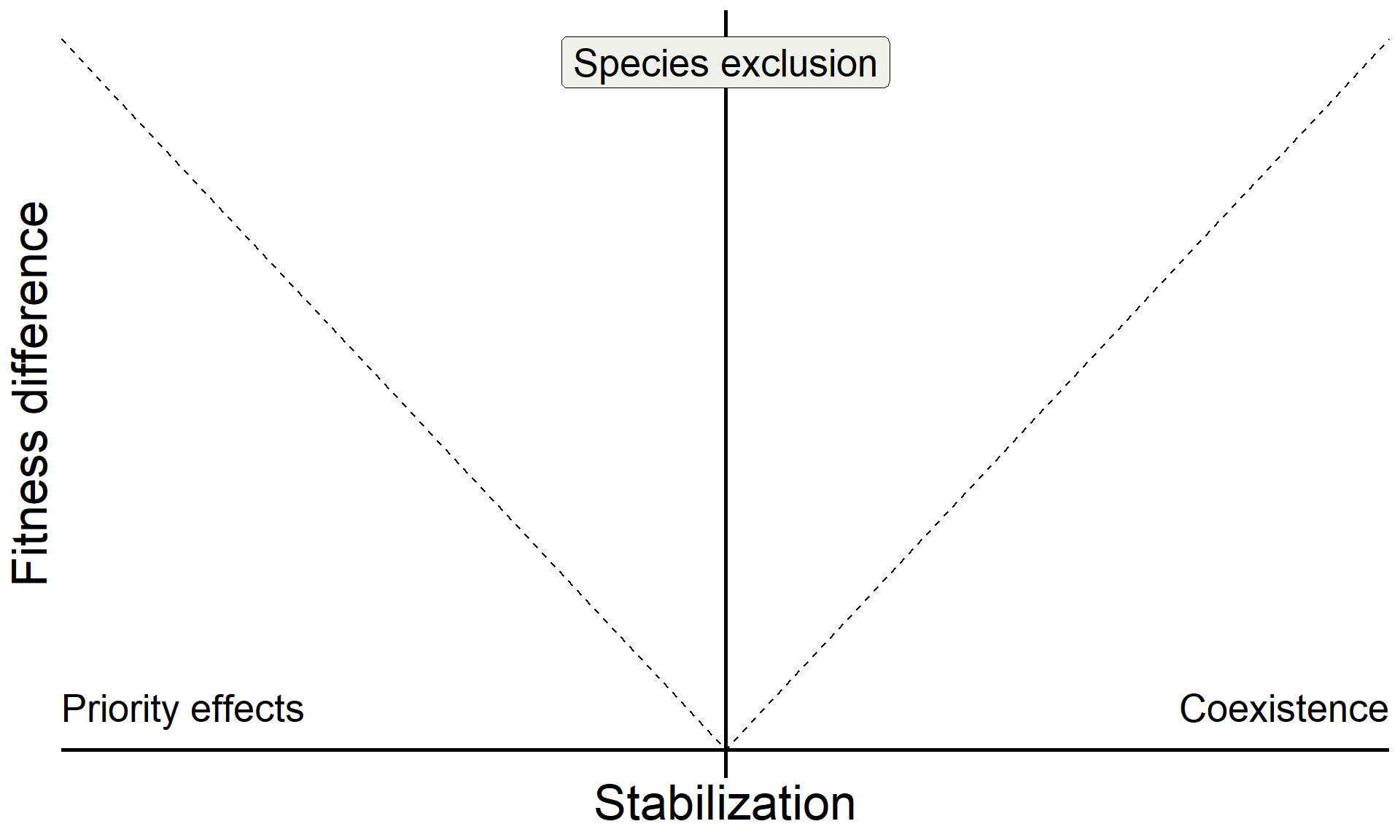
Code
metrics <- function(params) {
IS <- with(as.list(params), {m1A - m2A - m1B + m2B})
FD <- with(as.list(params), {(1/2)*(m1A+m2A) - (1/2)*(m1B+m2B)})
SD <- (-1/2)*IS
return(c(IS = IS, SD = SD, FD = FD))
}
coex_metrics <- metrics(params_coex)
base_coex <-
base +
inset_element({panel_freq_coex + theme(legend.position = 'none',
axis.text = element_blank(),
axis.ticks = element_blank(),
axis.title = element_blank(),
plot.background = element_rect(color = "grey"))},
0.75,0.15,1,0.5)
base_coex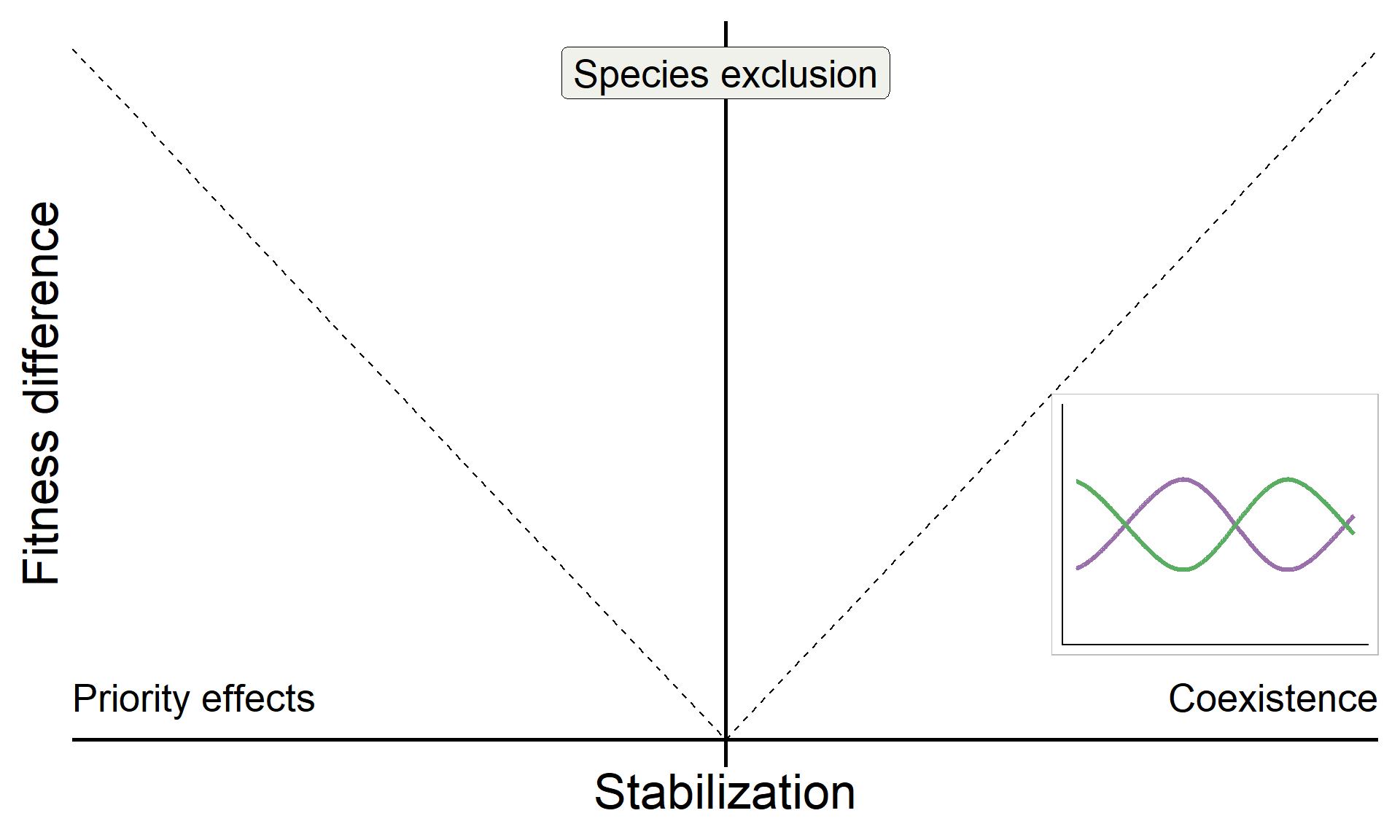
Do microbes generate fitness differences in nature?
Sedgwick Reserve (unceded territory of Chumash people)

Code
# Example point, using UR_FE values
base +
annotate("pointrange", x = 0.275, y = 0.0591, ymin = 0.0591-0.307,ymax = 0.0591+0.307, shape = 21, stroke = 1.5, size = 1, linewidth = 0.1, fill = "#4477aa") +
annotate("pointrange", x = 0.275, y = 0.0591, xmin = 0.275-0.152,xmax = 0.275+0.152, shape = 21, fill = "#4477aa", linewidth = 0.1, size = 1, stroke = 1.5) +
annotate("text", x = 0.275 + 0.3, y = 0.0591 + 0.15, color = "#4477aa", label = "Festuca - Plantago", size = 8, fontface = "italic") +
theme(axis.title = element_blank())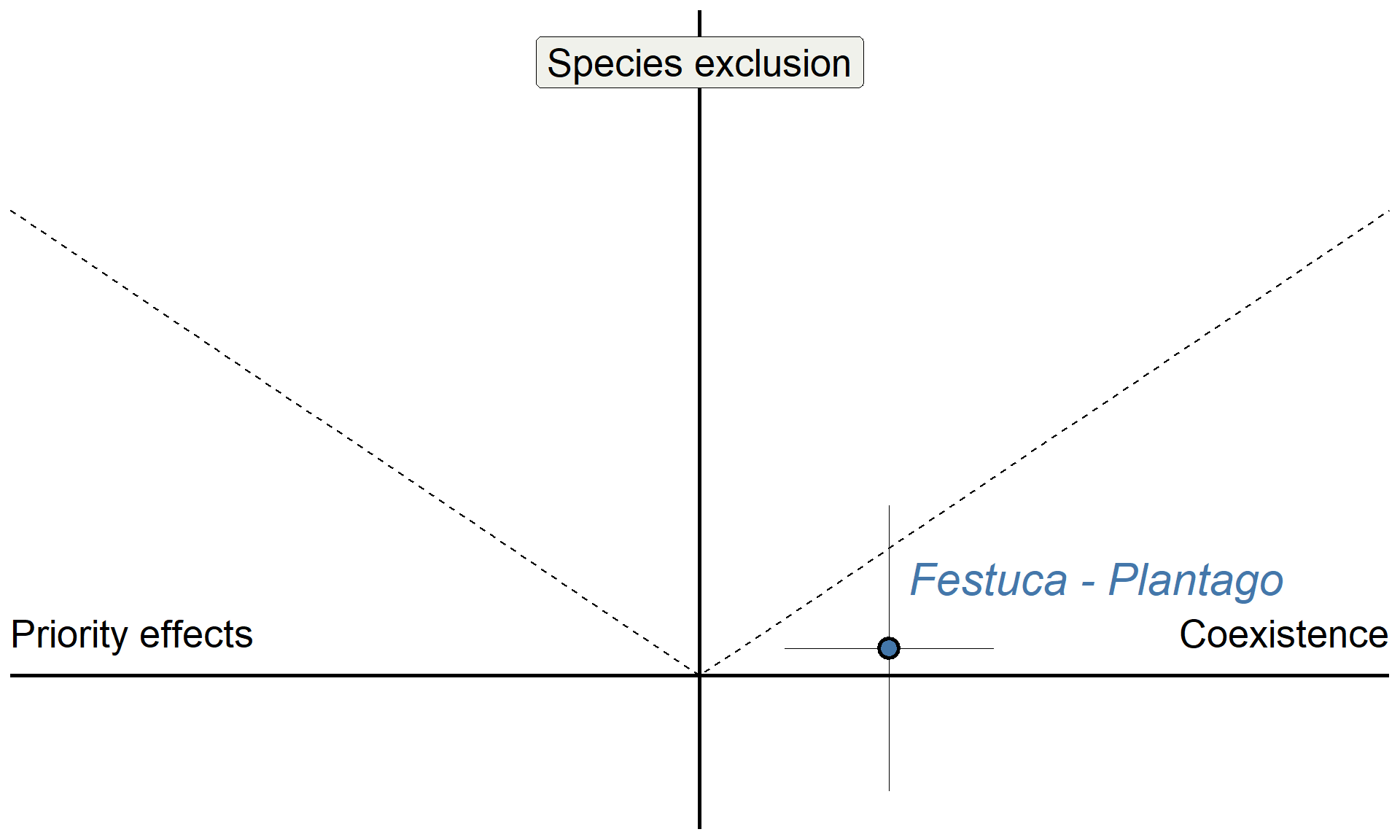
Code
# Dataset downloaded from Figshare:
# curl::curl_download("https://datadryad.org/downloads/file_stream/367188", destfile = "../data/kandlikar2021-supplement.csv")
biomass_wide <-
read_csv("../data/kandlikar2021-supplement.csv") |>
mutate(log_agb = log(abg_dry_g)) |>
mutate(source_soil = ifelse(source_soil == "aastr", "str", source_soil),
source_soil = ifelse(source_soil == "abfld", "fld", source_soil),
pair = paste0(source_soil, "_", focal_species)) |>
select(replicate, pair, log_agb) |>
pivot_wider(names_from = pair, values_from = log_agb) |>
filter(!is.na(replicate))
# Calculate the Stabilization between each species pair ----
# Recall that following the definition of the m terms in Bever 1997,
# and the analysis of this model in Kandlikar 2019,
# stablization = -0.5*(log(m1A) + log(m2B) - log(m1B) - log(m2A))
# Here is a function that does this calculation
# (recall that after the data reshaping above,
# each column represents a given value of log(m1A))
calculate_stabilization <- function(df) {
df |>
# In df, each row is one repilcate/rack, and each column
# represents the growth of one species in one soil type.
# e.g. "FE_ACWR" is the growth of FE in ACWR-cultivated soil.
mutate(AC_FE = -0.5*(AC_ACWR - AC_FEMI - FE_ACWR + FE_FEMI),
AC_HO = -0.5*(AC_ACWR - AC_HOMU - HO_ACWR + HO_HOMU),
AC_SA = -0.5*(AC_ACWR - AC_SACO - SA_ACWR + SA_SACO),
AC_PL = -0.5*(AC_ACWR - AC_PLER - PL_ACWR + PL_PLER),
AC_UR = -0.5*(AC_ACWR - AC_URLI - UR_ACWR + UR_URLI),
FE_HO = -0.5*(FE_FEMI - FE_HOMU - HO_FEMI + HO_HOMU),
FE_SA = -0.5*(FE_FEMI - FE_SACO - SA_FEMI + SA_SACO),
FE_PL = -0.5*(FE_FEMI - FE_PLER - PL_FEMI + PL_PLER),
FE_UR = -0.5*(FE_FEMI - FE_URLI - UR_FEMI + UR_URLI),
HO_PL = -0.5*(HO_HOMU - HO_PLER - PL_HOMU + PL_PLER),
HO_SA = -0.5*(HO_HOMU - HO_SACO - SA_HOMU + SA_SACO),
HO_UR = -0.5*(HO_HOMU - HO_URLI - UR_HOMU + UR_URLI),
SA_PL = -0.5*(SA_SACO - SA_PLER - PL_SACO + PL_PLER),
SA_UR = -0.5*(SA_SACO - SA_URLI - UR_SACO + UR_URLI),
PL_UR = -0.5*(PL_PLER - PL_URLI - UR_PLER + UR_URLI)) |>
select(replicate, AC_FE:PL_UR) |>
gather(pair, stabilization, AC_FE:PL_UR)
}
stabilization_values <- calculate_stabilization(biomass_wide)
# Seven values are NA; we can omit these
stabilization_values <- stabilization_values |>
filter(!(is.na(stabilization)))
# Calculating the Fitness difference between each species pair ----
# Similarly, we can now calculate the fitness difference between
# each pair. Recall that FD = 0.5*(log(m1A)+log(m1B)-log(m2A)-log(m2B))
# But recall that here, IT IS IMPORTANT THAT
# m1A = (m1_soilA - m1_fieldSoil)!
# The following function does this calculation:
calculate_fitdiffs <- function(df) {
df |>
mutate(AC_FE = 0.5*((AC_ACWR-fld_ACWR) + (FE_ACWR-fld_ACWR) - (AC_FEMI-fld_FEMI) - (FE_FEMI-fld_FEMI)),
AC_HO = 0.5*((AC_ACWR-fld_ACWR) + (HO_ACWR-fld_ACWR) - (AC_HOMU-fld_HOMU) - (HO_HOMU-fld_HOMU)),
AC_SA = 0.5*((AC_ACWR-fld_ACWR) + (SA_ACWR-fld_ACWR) - (AC_SACO-fld_SACO) - (SA_SACO-fld_SACO)),
AC_PL = 0.5*((AC_ACWR-fld_ACWR) + (PL_ACWR-fld_ACWR) - (AC_PLER-fld_PLER) - (PL_PLER-fld_PLER)),
AC_UR = 0.5*((AC_ACWR-fld_ACWR) + (UR_ACWR-fld_ACWR) - (AC_URLI-fld_URLI) - (UR_URLI-fld_URLI)),
FE_HO = 0.5*((FE_FEMI-fld_FEMI) + (HO_FEMI-fld_FEMI) - (FE_HOMU-fld_HOMU) - (HO_HOMU-fld_HOMU)),
FE_SA = 0.5*((FE_FEMI-fld_FEMI) + (SA_FEMI-fld_FEMI) - (FE_SACO-fld_SACO) - (SA_SACO-fld_SACO)),
FE_PL = 0.5*((FE_FEMI-fld_FEMI) + (PL_FEMI-fld_FEMI) - (FE_PLER-fld_PLER) - (PL_PLER-fld_PLER)),
FE_UR = 0.5*((FE_FEMI-fld_FEMI) + (UR_FEMI-fld_FEMI) - (FE_URLI-fld_URLI) - (UR_URLI-fld_URLI)),
HO_PL = 0.5*((HO_HOMU-fld_HOMU) + (PL_HOMU-fld_HOMU) - (HO_PLER-fld_PLER) - (PL_PLER-fld_PLER)),
HO_SA = 0.5*((HO_HOMU-fld_HOMU) + (SA_HOMU-fld_HOMU) - (HO_SACO-fld_SACO) - (SA_SACO-fld_SACO)),
HO_UR = 0.5*((HO_HOMU-fld_HOMU) + (UR_HOMU-fld_HOMU) - (HO_URLI-fld_URLI) - (UR_URLI-fld_URLI)),
SA_PL = 0.5*((SA_SACO-fld_SACO) + (PL_SACO-fld_SACO) - (SA_PLER-fld_PLER) - (PL_PLER-fld_PLER)),
SA_UR = 0.5*((SA_SACO-fld_SACO) + (UR_SACO-fld_SACO) - (SA_URLI-fld_URLI) - (UR_URLI-fld_URLI)),
PL_UR = 0.5*((PL_PLER-fld_PLER) + (UR_PLER-fld_PLER) - (PL_URLI-fld_URLI) - (UR_URLI-fld_URLI))) |>
select(replicate, AC_FE:PL_UR) |>
gather(pair, fitdiff_fld, AC_FE:PL_UR)
}
fd_values <- calculate_fitdiffs(biomass_wide)
# Twenty-two values are NA; let's omit these.
fd_values <- fd_values |> filter(!(is.na(fitdiff_fld)))
# Now, generate statistical summaries of SD and FD
stabiliation_summary <- stabilization_values |> group_by(pair) |>
summarize(mean_sd = mean(stabilization),
sem_sd = sd(stabilization)/sqrt(n()),
n_sd = n())
fitdiff_summary <- fd_values |> group_by(pair) |>
summarize(mean_fd = mean(fitdiff_fld),
sem_fd = sd(fitdiff_fld)/sqrt(n()),
n_fd = n())
# Combine the two separate data frames.
sd_fd_summary <- left_join(stabiliation_summary, fitdiff_summary)
# Some of the FDs are negative, let's flip these to be positive
# and also flip the label so that the first species in the name
# is always the fitness superior.
sd_fd_summary <- sd_fd_summary |>
# if mean_fd is < 0, the following command gets the absolute
# value and also flips around the species code so that
# the fitness superior is always the first species in the code
mutate(pair = ifelse(mean_fd < 0,
paste0(str_extract(pair, "..$"),
"_",
str_extract(pair, "^..")),
pair),
mean_fd = abs(mean_fd))
sd_fd_summary <-
sd_fd_summary |>
mutate(
# outcome = ifelse(mean_fd - 2*sem_fd >
# mean_sd + 2*sem_sd, "exclusion", "neutral"),
outcome2 = ifelse(mean_fd > mean_sd, "exclusion", "coexistence"),
outcome2 = ifelse(mean_sd < 0, "exclusion or priority effect", outcome2)
)
base +
geom_pointrange(data = sd_fd_summary,
aes(x = mean_sd, y = mean_fd,
ymin = mean_fd-sem_fd,
ymax = mean_fd+sem_fd, fill = outcome2), size = 1, linewidth = 0.1,
shape = 21, stroke = 1.5) +
geom_pointrange(data = sd_fd_summary,
aes(x = mean_sd, y = mean_fd,
xmin = mean_sd-sem_sd,
xmax = mean_sd+sem_sd, fill = outcome2), size = 1, linewidth = 0.1, shape = 21, stroke = 1.5) +
scale_fill_manual(values = c("#4477aa", "#ee6677", "#ccbb44")) +
theme(legend.position = "none")+
theme(axis.title = element_blank())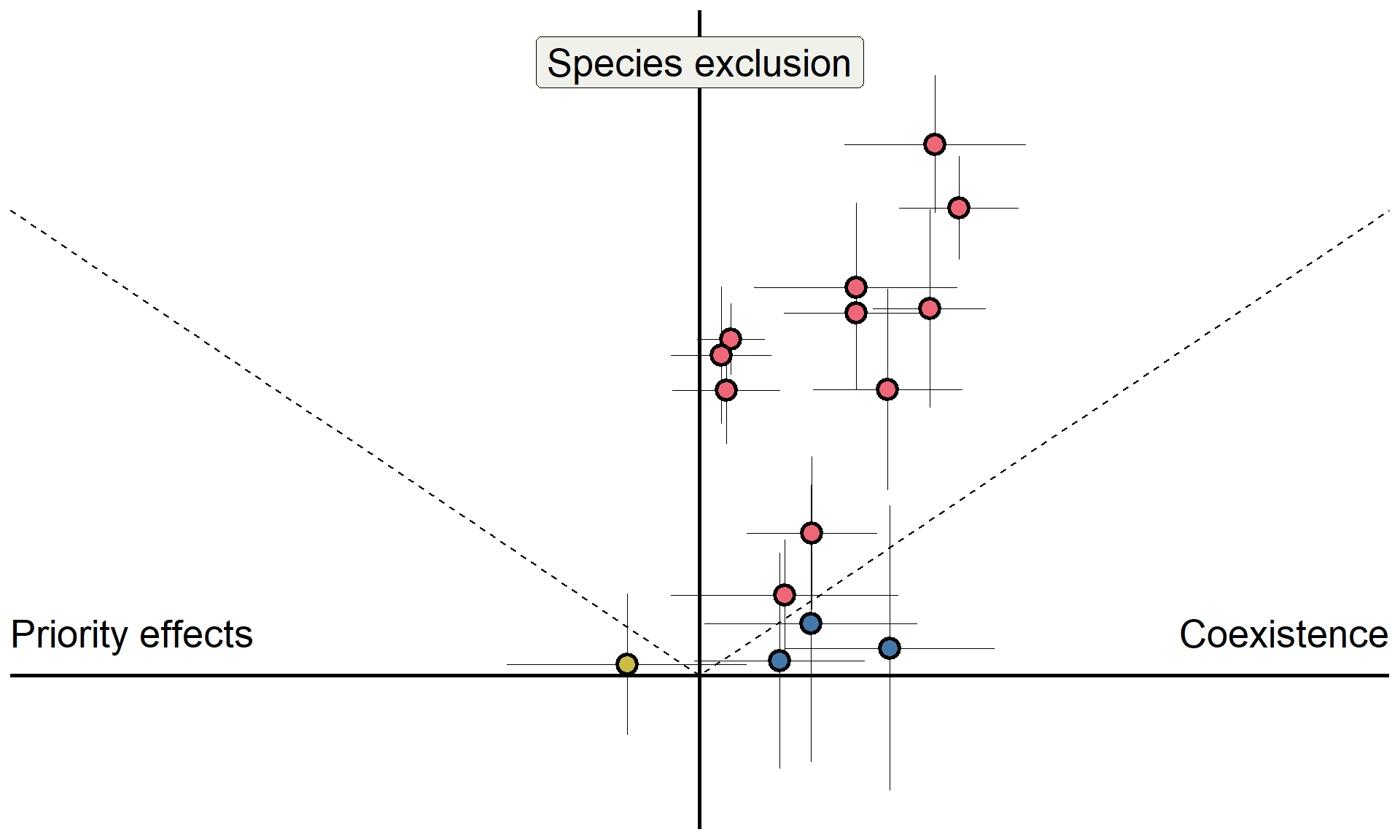
Detailed in Kandlikar et al. (2021)
Do microbes generate fitness differences in nature?
In California grasslands, microbes tend to drive stronger fitness differences than stabilization.
Can we answer this question more generally?

Meta-analysis overview
Data for 518 pairwise comparisons of stabilization and fitness differences (results shown for 72 pairs here)
New sampling approach for quantifying uncertainty in coexistence outcomes
Results in Yan, Levine, and Kandlikar (2022); Analyses available on Zenodo
Do microbes generate fitness differences in nature?
- Fitness differences represent a dominant avenue through which microbes control plant coexistence
Microbially-mediated intransitivity in pine-oak forests in Spain, Pajares-Murgó et al. (2024)
Today’s talk
- Ecological theory for microbial effects on plant coexistence
2. Microbial effects on plant communities in variable environments
Today’s talk
- Ecological theory for microbial effects on plant coexistence
2. Microbial effects on plant communities in variable environments
Coexistence in fragmented tropical forests
Woody encroachment in longleaf pine understories
Plant eco-evolutionary dynamics under drought
Hassan District, Karnataka, India
Gradients in soil moisture from forest edge to interior
Gradients in soil moisture from forest edge to interior
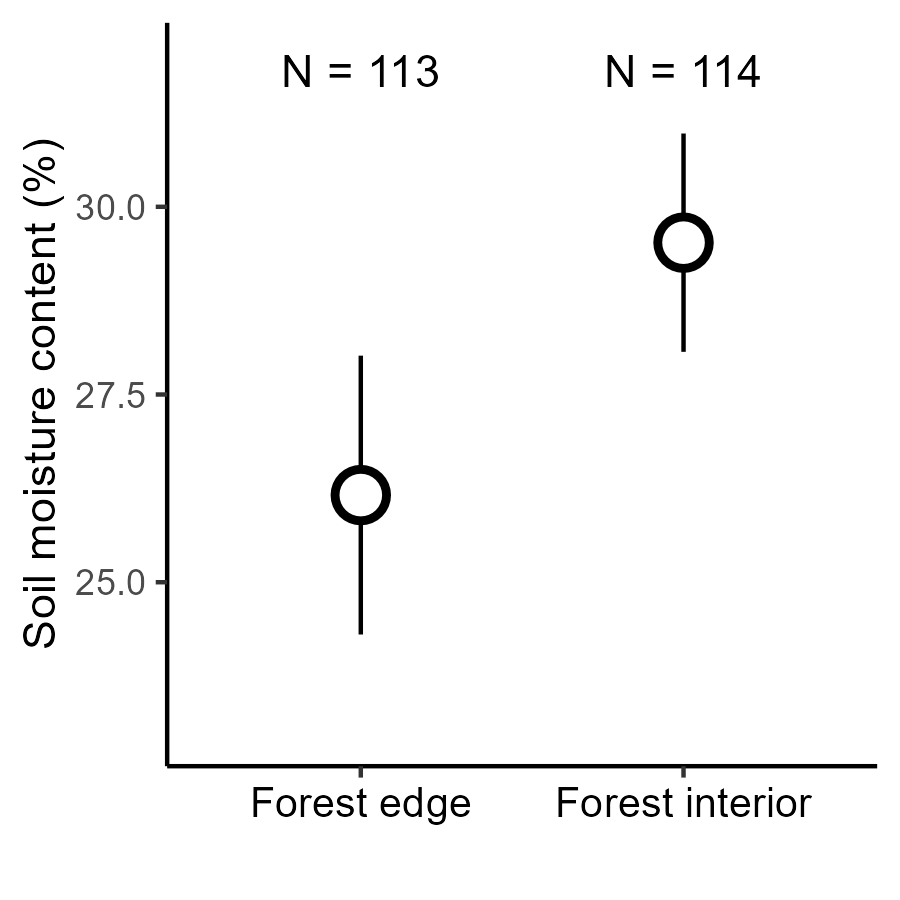
Gradients in soil moisture from forest edge to interior

Gradients in soil moisture from forest edge to interior

Detailed in Krishnadas et al. (2018)
Can variable plant–soil feedback explain this result?
Quantify pairwise plant–soil feedback under high and low water conditions in a shadehouse
Exaggerated microbially mediated fitness differences in dry soils could contribute to erosion of diversity at fragment edges.
Analyses ongoing
Applications of basic ecology to habitat management

PhD student
Do soil microbes contribute to invasion of coffee into forest edges?
Do soil microbes in abandoned coffee orchards affect restoration?
Applications of basic ecology to habitat management

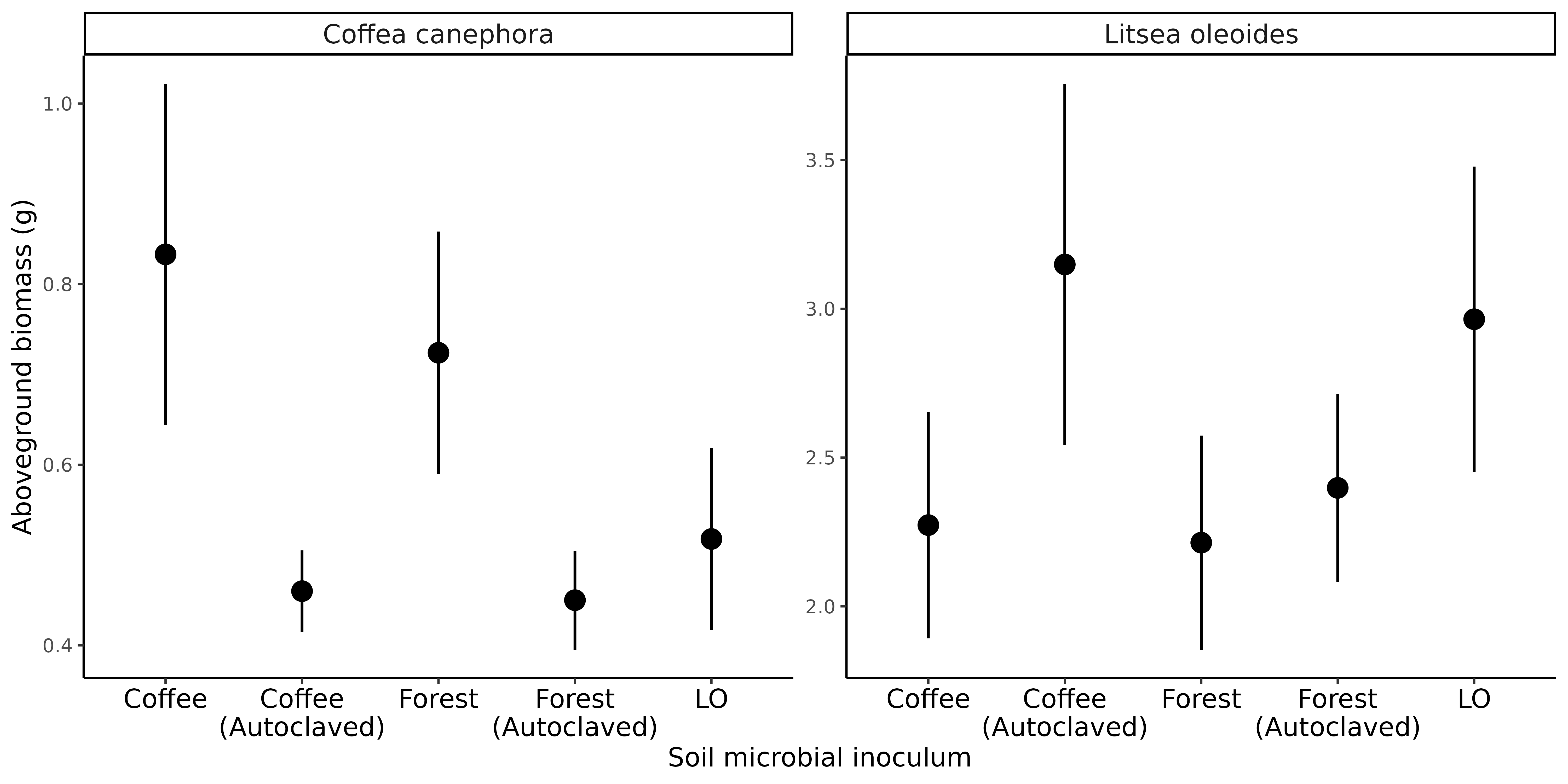
Extremely preliminary analyses!
Coffee growth enhanced by forest soil microbes
Microbes in coffee soil suppress growth of native seedlings
Today’s talk
- Ecological theory for microbial effects on plant coexistence
2. Microbial effects on plant communities in variable environments
Coexistence in fragmented tropical forests
Woody encroachment in longleaf pine understories
Plant eco-evolutionary dynamics under drought


Longleaf pine savanna understory at Palustris Experimental Forest
Across ecosystems, grasses tend to experience negative feedback
Code
# curl::curl_download("https://datadryad.org/downloads/file_stream/2789266", destfile = "../data/jiang2024-dataset.csv")
jiang_dat <- read_csv("../data/jiang2024-dataset.csv")
# jiang_metamod <- metafor::rma.mv(rr, var, data = jiang_dat, mods = ~ Life.form-1)
# saveRDS(jiang_metamod, "../data/jiang-metamod.rds")
metamod <- readRDS("../data/jiang-metamod.rds")
metamod_s <- broom::tidy(metamod)
jiang_dat |>
ggplot(aes(x = rr, y = Life.form, color = Life.form)) +
ggbeeswarm::geom_quasirandom(shape = 21) +
scale_color_manual(values = c("#ccbb44","#228833")) +
annotate("point",
x = metamod_s$estimate[1], y = 1, size = 3, shape = 21, stroke = 1.5) +
annotate("point",
x = metamod_s$estimate[2], y = 2, size = 3, shape = 21, stroke = 1.5) +
ylab("") +
xlab("Growth in self vs. growth in non-self") +
xlim(-2.5,2.5)+
geom_vline(xintercept = 0, linetype = 'dashed') +
theme(legend.position = "none",
axis.text.y = element_text(size = 12, color = 'black'))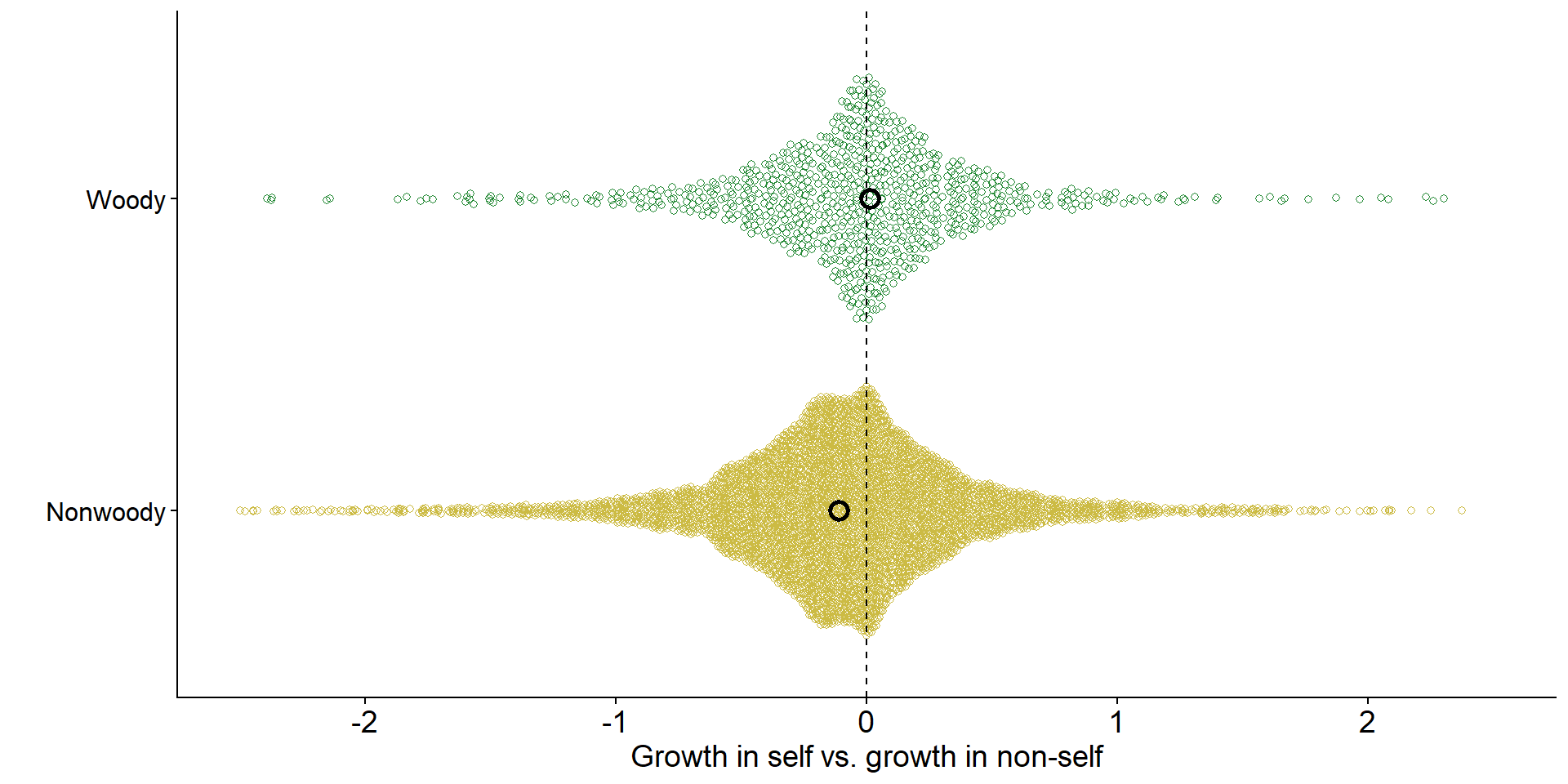
Data from Jiang et al. (2024)
Do soil microbes play a role in driving shrubification under fire suppression?
What are the direct impacts of fire on soil microbial dynamics?
Does the history and severity of fire mediate the impact of soil microbes on plant performance and coexistence?

Postdoc
Approach
Field experiment: evaluate fire effects on soil communities
Hoophouse experiment: evaluate shrub and grass growth in shrub- and grass-conditioned soils with different burn histories
Mathematical modeling: experimentally-informed dynamics of plant community dynamics




Plot set to burn next week
How does burn severity affect biotic and abiotic soil properties?
What are the dynamics of recovery?
Mathematical modeling of plant dynamics
- Goal: translate results from short-term experiments into long-term projections of community dynamics
Today’s talk
- Ecological theory for microbial effects on plant coexistence
2. Microbial effects on plant communities in variable environments
Coexistence in fragmented tropical forests
Woody encroachment in longleaf pine understories
Plant eco-evolutionary dynamics under drought
Past work: Soil microbial legacies mediate plant response to drought across generations
Code
# curl::curl_download("https://datadryad.org/downloads/file_stream/50543", destfile = "../data/lau2012-dataset.csv")
lau_dat <- read_csv("../data/lau2012-dataset.csv")
lau_dat |>
group_by(Contemp_Water, Plant_History, Microbe_History) |>
filter(!is.na(Fruit_number)) |>
summarize(mean_fruit = mean(Fruit_number),
sd_fruit = sd(Fruit_number),
nreps = n(),
sem = sqrt(sd_fruit/(nreps-1))) |>
mutate(Microbe_History =
ifelse(Microbe_History == "Dry microbes", "Dry-adapted\nmicrobes", "Wet-adapted\nmicrobes")) |>
filter(Plant_History == "Wet") |> # Only show wet history plants for simplicity
ggplot(aes(x = Contemp_Water, y = mean_fruit,
ymin = mean_fruit-sem,
ymax = mean_fruit+sem,
fill = Microbe_History)) +
geom_pointrange(shape = 21,
size = 1.25,
linewidth = 1,
position = position_dodge(width = 0.25)) +
scale_fill_manual(name = "",
values = c("#cfe2f3ff", "#0b5394ff")) +
scale_y_continuous(#limits = c(0,6),
name = "Fruit number") +
xlab("Contemporary watering") +
# facet_wrap(.~Plant_History, scales = "free") +f
theme(axis.text = element_text(size = 12, color = "black"),
legend.text = element_text(size = 12),
legend.position = "inside",
legend.position.inside = c(0.8,0.2),
legend.background = element_rect(color = "black"))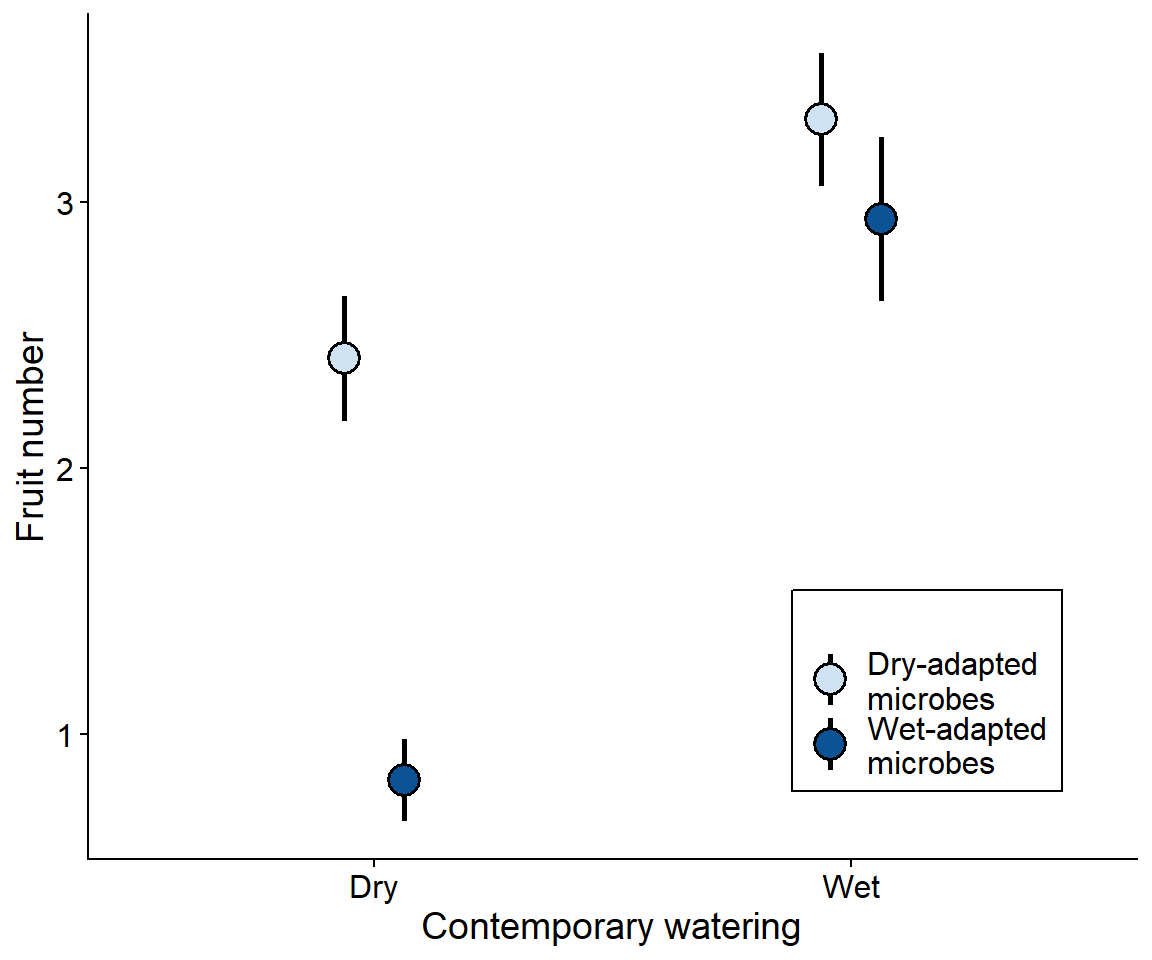
Brassica rapa’s tolerance of dought is enhanced by growing with a drought-adapted soil microbiome Data from Lau and Lennon (2012)
Ongoing/future work: What is the mechanistic basis, and how commonly do microbes shape transgenerational plant dynamics under abiotic stress?


Postbac Scholar
Currently in the second generation of experimental drought, with additional plants grown to evaluate immediate maternal effects on plant functional traits
Drought imposes a fitness cost to B. rapa

Extremely preliminary analyses!
Drought imposes a fitness cost to B. rapa
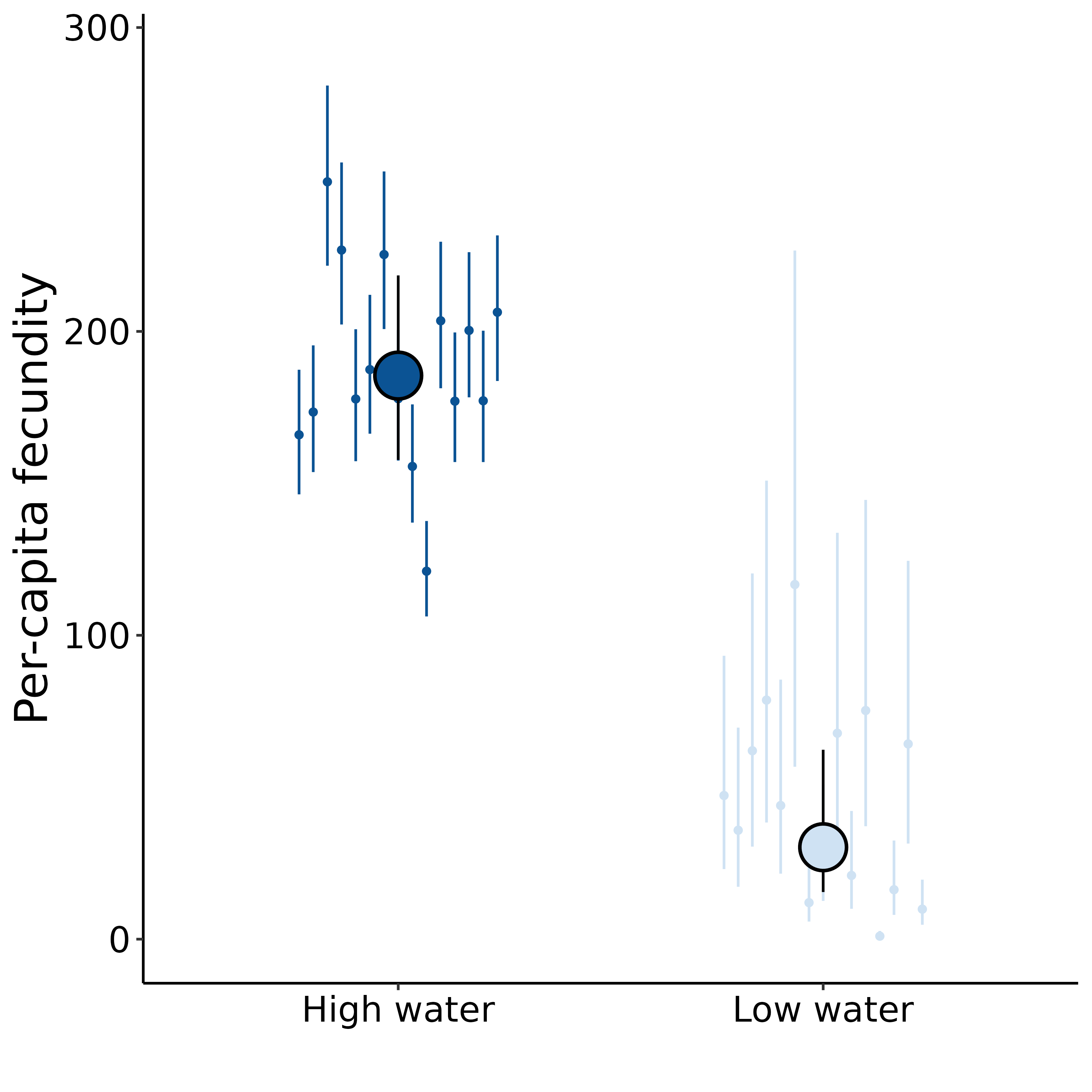
Extremely preliminary analyses!
Drought imposes a fitness cost to B. rapa

Evidence of genetic variation in drought tolerance
Extremely preliminary analyses!
Currently in the second generation of experimental drought, with additional plants grown to evaluate immediate maternal effects on plant functional traits
Early indication of maternal effects
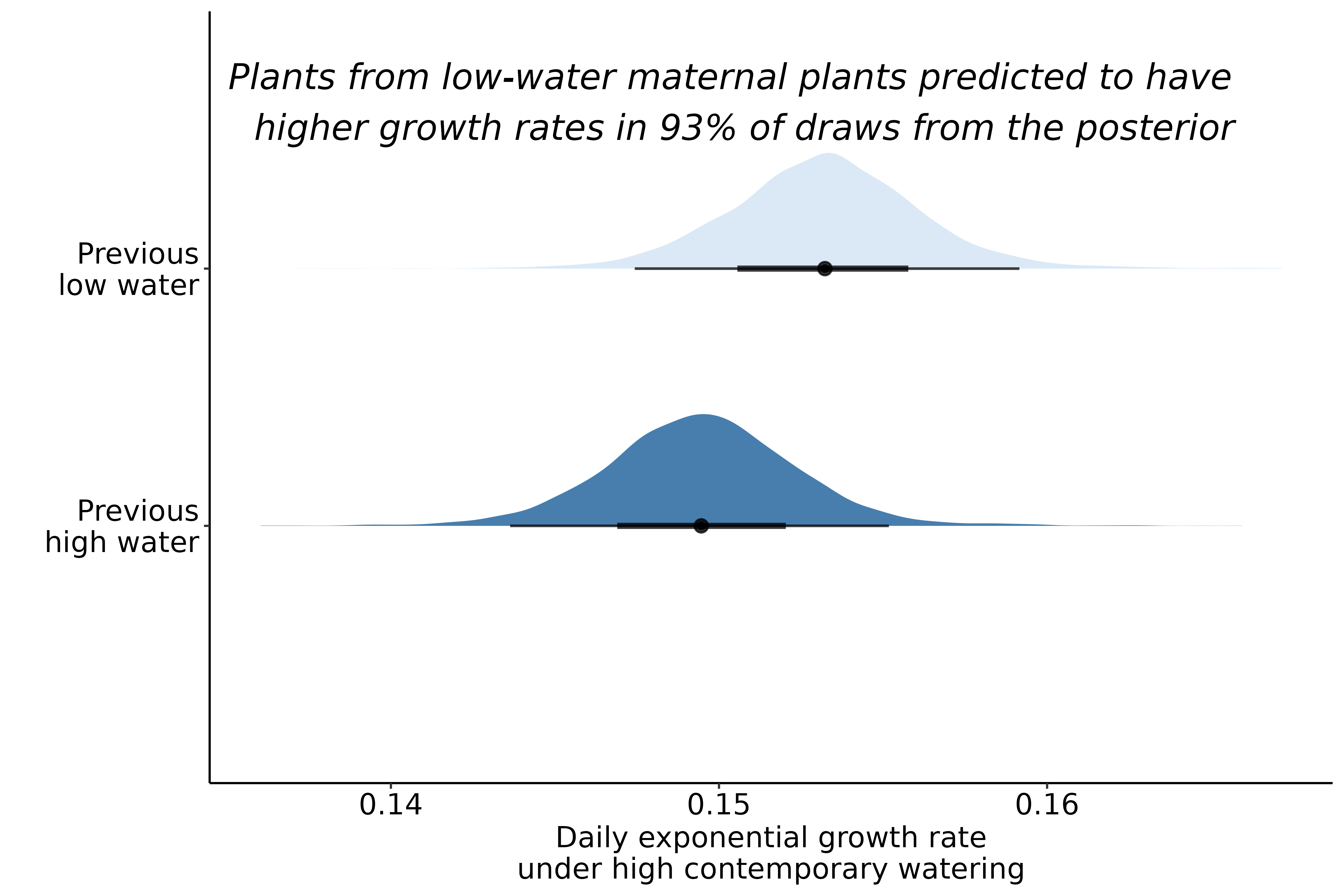
Extremely preliminary analyses!

Other projects in the lab

PhD student
How does nutrient enrichment structure plant–mycorrhizal symbiosis?

PhD student
Citizen science for invasion ecology
Thank you!
slides: https://talks.gklab.org/tulane-25
contact: gkandlikar@lsu.edu