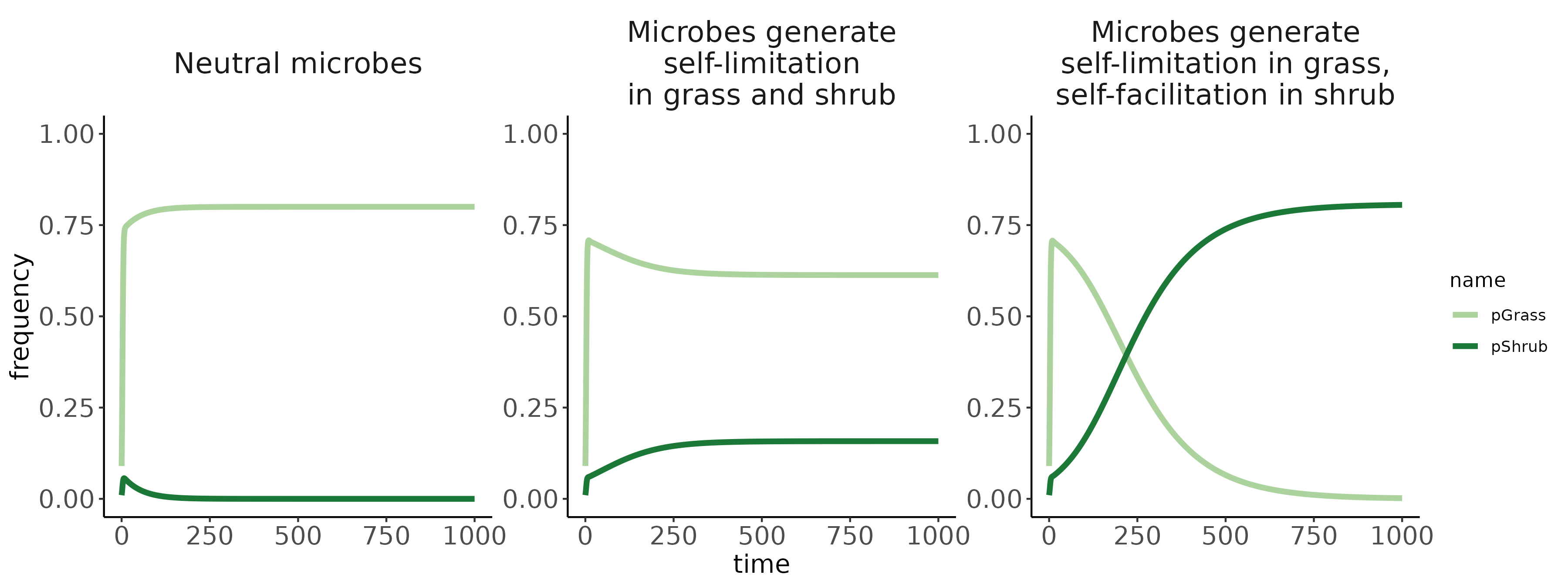
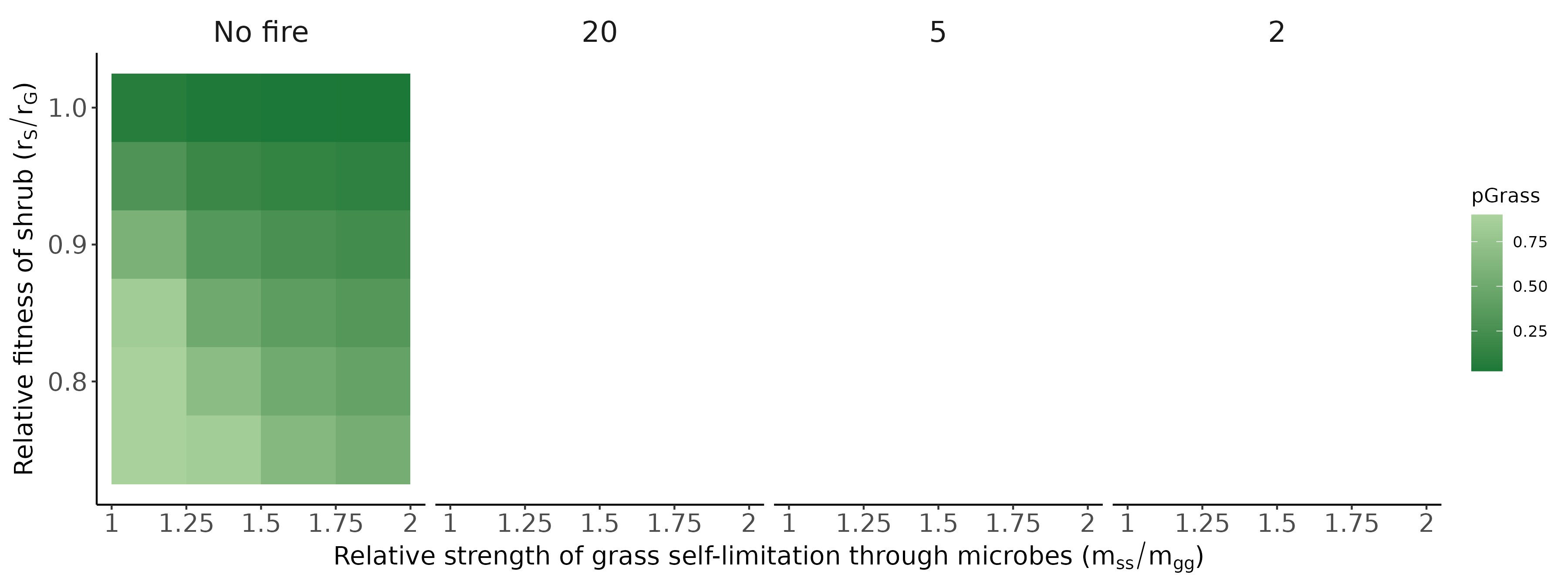
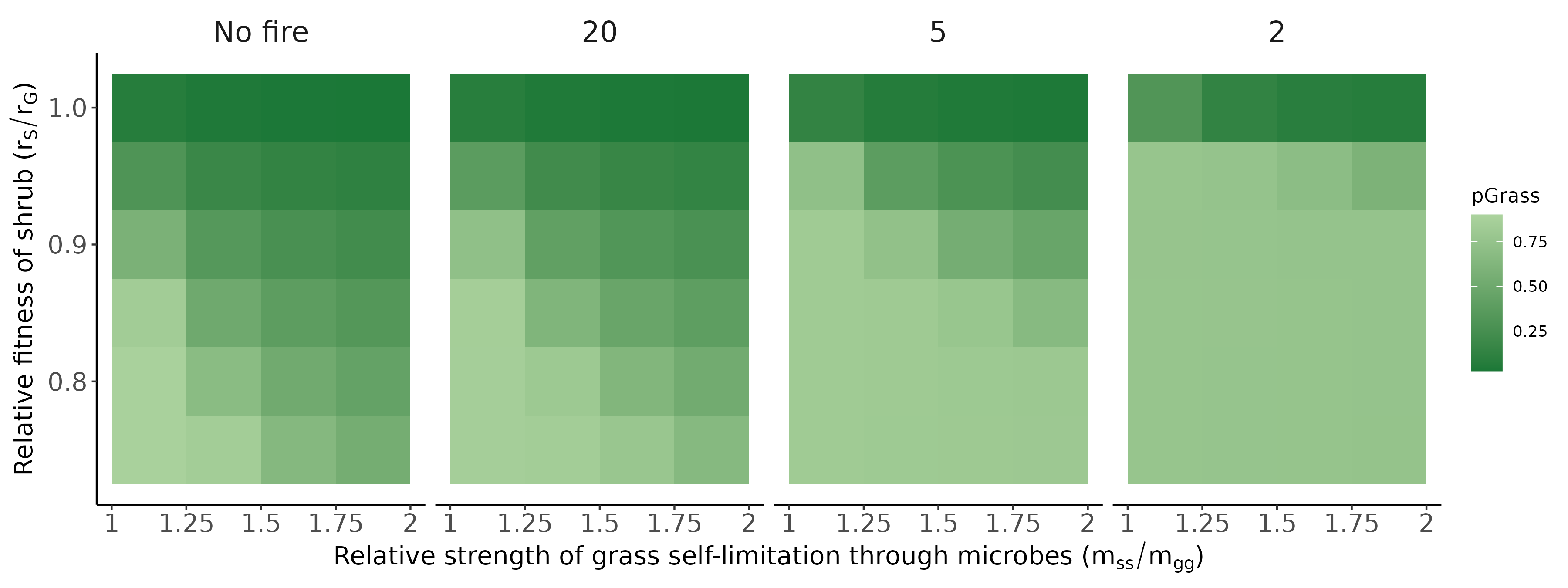
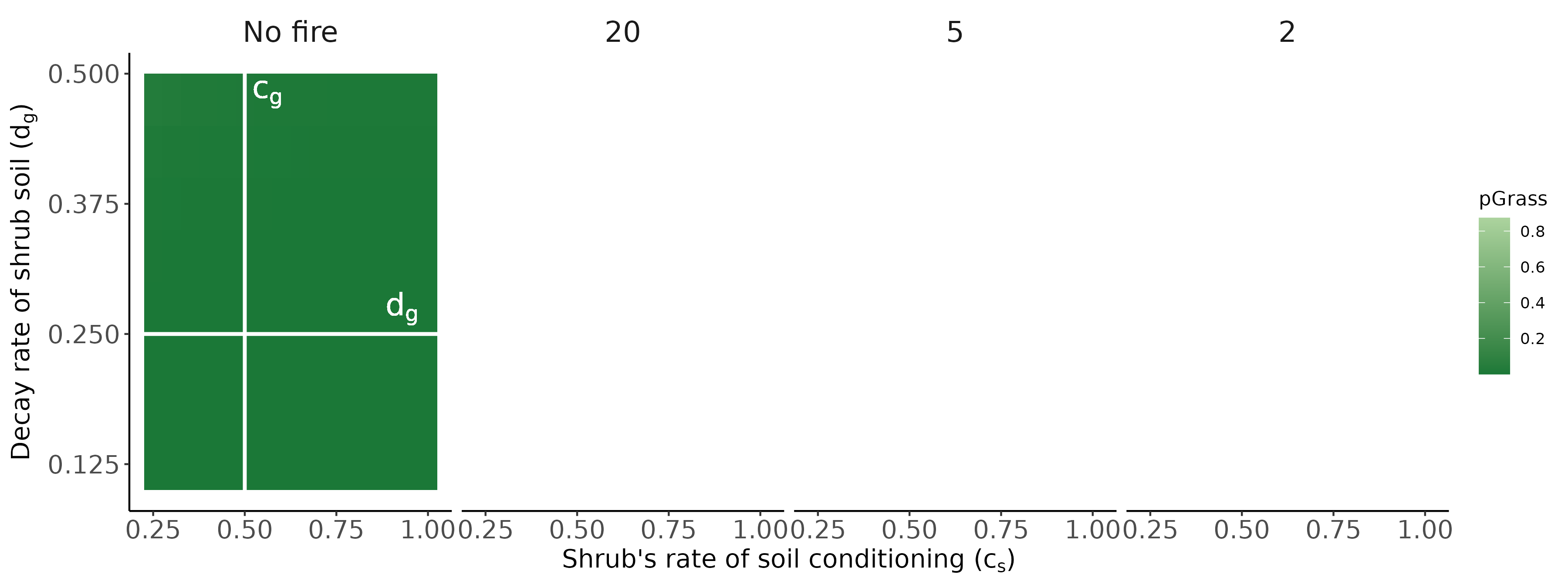
Bennett, Jonathan A, Hafiz Maherali, Kurt O Reinhart, Ylva Lekberg, Miranda M Hart, and John Klironomos. 2017. “Plant-Soil Feedbacks and Mycorrhizal Type Influence Temperate Forest Population Dynamics.” Science 355 (6321): 181–84.
Jiang, Feng, Jonathan A Bennett, Kerri M Crawford, Johannes Heinze, Xucai Pu, Ao Luo, and Zhiheng Wang. 2024. “Global Patterns and Drivers of Plant–Soil Microbe Interactions.” Ecology Letters 27 (1): e14364.
Ke, Po-Ju, Gaurav Kandlikar, Suzanne Xianran Ou, Gen-Chang Hsu, Joe Wan, and Meghna Krishnadas. 2024. “Time Will Tell: The Temporal and Demographic Contexts of Plant-Soil Microbe Interactions.”
Ke, Po-Ju, and Jonathan M Levine. 2021. “The Temporal Dimension of Plant-Soil Microbe Interactions: Mechanisms Promoting Feedback Between Generations.” The American Naturalist 198 (3): E80–94.